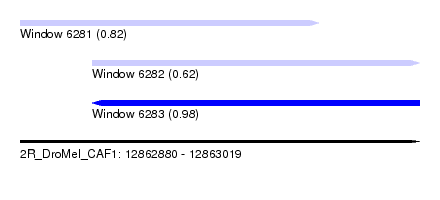
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,862,880 – 12,863,019 |
| Length | 139 |
| Max. P | 0.984593 |

| Location | 12,862,880 – 12,862,984 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 99.04 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12862880 104 + 20766785 UGCUUUUCCACAAUGGCAUGCCAGAACGCUAGACGACA-GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCA ............((((((((((((...((((..(((.(-(((....)))))))..))))))))))...........(((((........)))))...)))))).. ( -30.80) >DroSim_CAF1 218189 104 + 1 UGCUUUUCCACAAUGGCAUGCCAGAACGCUAGACGACA-GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCA ............((((((((((((...((((..(((.(-(((....)))))))..))))))))))...........(((((........)))))...)))))).. ( -30.80) >DroEre_CAF1 210386 105 + 1 UGCUUUUCCACAAUGGCAUGCCAGAACGCUAGACGACAGGCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCA ............((((((((((((...((((..(((..((((....)))))))..))))))))))...........(((((........)))))...)))))).. ( -29.80) >DroYak_CAF1 216258 104 + 1 UGCUUUUCCACAAUGGCAUGCCAGGACGCUAGACGACA-GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCA ............(((((((((((((.((((.......)-))).((((......)))).)))))))...........(((((........)))))...)))))).. ( -32.40) >consensus UGCUUUUCCACAAUGGCAUGCCAGAACGCUAGACGACA_GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCA ............((((((((((((...((((..(((...(((....))).)))..))))))))))...........(((((........)))))...)))))).. (-29.52 = -29.53 + 0.00)



| Location | 12,862,905 – 12,863,019 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12862905 114 + 20766785 ACGCUAGACGACA-GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCAUGCCACCCGAAAAAGGAUAAAAAAA-----AAGAAAAGCA .((((.......)-)))......(((((..((((.(((((...........(((((........)))))...)))))....))))..))))).............-----.......... ( -24.14) >DroEre_CAF1 210411 111 + 1 ACGCUAGACGACAGGCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCAUGCCACCCGAAAAAGGAUGAAAAAAAAGGGA--------- .((((........)))).((((.(((((..((((.(((((...........(((((........)))))...)))))....))))..)))))...))))............--------- ( -27.24) >DroYak_CAF1 216283 119 + 1 ACGCUAGACGACA-GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCAUGCCACCCGAAAAAGGAUAAAAAAAAAAGAAAAACGAGUA .((((.......)-)))......(((((..((((.(((((...........(((((........)))))...)))))....))))..)))))............................ ( -24.14) >consensus ACGCUAGACGACA_GCGACAUUGUUUCGAAUGGCCUGGCAAAAGAAGAAAAAUGGCUCUGAAAUGCCAUCACUGCCAUCAUGCCACCCGAAAAAGGAUAAAAAAAAA_G_AA_A__AG_A .(((..........)))......(((((..((((.(((((...........(((((........)))))...)))))....))))..)))))............................ (-22.34 = -22.34 + -0.00)


| Location | 12,862,905 – 12,863,019 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12862905 114 - 20766785 UGCUUUUCUU-----UUUUUUUAUCCUUUUUCGGGUGGCAUGAUGGCAGUGAUGGCAUUUCAGAGCCAUUUUUCUUCUUUUGCCAGGCCAUUCGAAACAAUGUCGC-UGUCGUCUAGCGU ..........-----.............(((((((((((....((((((.((((((........)))))).........)))))).)))))))))))......(((-((.....))))). ( -34.60) >DroEre_CAF1 210411 111 - 1 ---------UCCCUUUUUUUUCAUCCUUUUUCGGGUGGCAUGAUGGCAGUGAUGGCAUUUCAGAGCCAUUUUUCUUCUUUUGCCAGGCCAUUCGAAACAAUGUCGCCUGUCGUCUAGCGU ---------...................(((((((((((....((((((.((((((........)))))).........)))))).)))))))))))......(((..(....)..))). ( -32.40) >DroYak_CAF1 216283 119 - 1 UACUCGUUUUUCUUUUUUUUUUAUCCUUUUUCGGGUGGCAUGAUGGCAGUGAUGGCAUUUCAGAGCCAUUUUUCUUCUUUUGCCAGGCCAUUCGAAACAAUGUCGC-UGUCGUCUAGCGU ............................(((((((((((....((((((.((((((........)))))).........)))))).)))))))))))......(((-((.....))))). ( -34.60) >consensus U_CU__U_UU_C_UUUUUUUUUAUCCUUUUUCGGGUGGCAUGAUGGCAGUGAUGGCAUUUCAGAGCCAUUUUUCUUCUUUUGCCAGGCCAUUCGAAACAAUGUCGC_UGUCGUCUAGCGU ............................(((((((((((....((((((.((((((........)))))).........)))))).)))))))))))......(((.((.....))))). (-30.80 = -30.80 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:34 2006