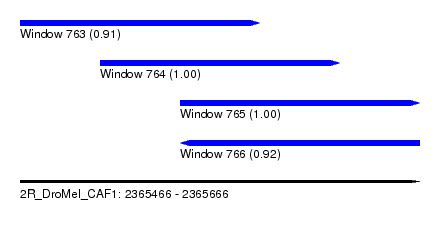
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,365,466 – 2,365,666 |
| Length | 200 |
| Max. P | 0.999445 |

| Location | 2,365,466 – 2,365,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.81 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2365466 120 + 20766785 CUCACAUUCGGGCUACCACAAAGCGACACAAUUUGAACUUUAACACCUUCGGUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC .........(((((.......))).......((((........(.(((..(((.((((....))))...)))..))).)...((((((((.....))))))))....)))).....)).. ( -23.80) >DroSec_CAF1 14979 115 + 1 -----ACUCGGGCUACCACAAAGCGACACAAUUUGAACUUUAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC -----....(((((.......))).......((((........(.((((((...((((....)))).)))....))).)...((((((((.....))))))))....)))).....)).. ( -20.30) >DroSim_CAF1 15095 120 + 1 CACUCACUCGGGCUACCACAAAGCGACACAAUUUGAACUUUAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC .........(((((.......))).......((((........(.((((((...((((....)))).)))....))).)...((((((((.....))))))))....)))).....)).. ( -20.30) >DroEre_CAF1 14619 113 + 1 -----ACUC-GGCUACCACAAAGCGACACA-UUUGAACUUUAACACCUUCAAUGACAGCCAGCUGUAUGACCUCCGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC -----....-((..........(((.(.((-((.(((..........)))))))((((....)))).........).)))..((((((((.....)))))))).............)).. ( -18.80) >DroYak_CAF1 14364 120 + 1 CACUCACUCGGGCUACCACAAAGCGAGACAAUUUGAACUUUAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC ..(((.((.((....))....)).)))....((((........(.((((((...((((....)))).)))....))).)...((((((((.....))))))))....))))......... ( -21.60) >consensus C_C_CACUCGGGCUACCACAAAGCGACACAAUUUGAACUUUAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUC .........(((((.......))).......((((........(.((((((...((((....)))).)))....))).)...((((((((.....))))))))....)))).....)).. (-18.60 = -18.84 + 0.24)

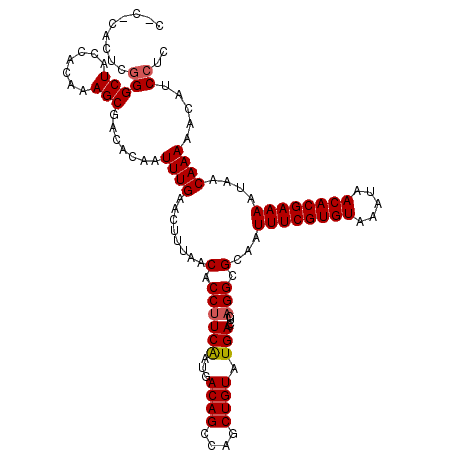
| Location | 2,365,506 – 2,365,626 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2365506 120 + 20766785 UAACACCUUCGGUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUUGAGAAUAAGGCUCCGACCUCUCGACCACGCCC ...((((...))))((((....)))).........((((...((((((((.....))))))))...............(((((.(((((.((.......))))))).)))))...)))). ( -36.40) >DroSec_CAF1 15014 120 + 1 UAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGACGAGAAGAAGGCUCCGACCUCUGGACCACGCCC ........(((...((((....)))).))).....((((...((((((((.....))))))))..............((((......)))).....((.((((.....)))))).)))). ( -29.10) >DroSim_CAF1 15135 120 + 1 UAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGAAGGCUCCGACCUCUGGACCACGCCC ........(((...((((....)))).))).....((((...((((((((.....))))))))...............((.((.(((((.((.......))))))).)).))...)))). ( -32.30) >DroEre_CAF1 14652 120 + 1 UAACACCUUCAAUGACAGCCAGCUGUAUGACCUCCGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGCAGAAGGCUCCGACCUCCGGACCACGCCC ........(((...((((....)))).))).....((((...((((((((.....))))))))...............(((.(.((((((((.....))).))))).))))....)))). ( -34.00) >DroYak_CAF1 14404 120 + 1 UAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGGAGGCUCCGACCUCUGGACCACGCCC ........(((...((((....)))).))).....((((...((((((((.....)))))))).....................((((.(((.(..(....)..).))).)))).)))). ( -33.20) >consensus UAACACCUUCAAUGACAGCCAGCUGUAUGACCUCAGGCGCAAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGAAGGCUCCGACCUCUGGACCACGCCC ........(((...((((....)))).))).....((((...((((((((.....))))))))...............((.((.(((((((.......)).))))).)).))...)))). (-29.54 = -29.62 + 0.08)

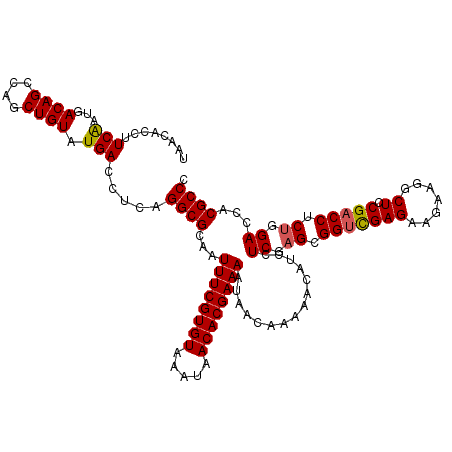
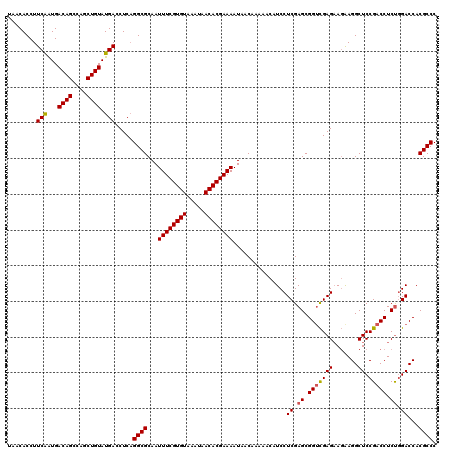
| Location | 2,365,546 – 2,365,666 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2365546 120 + 20766785 AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUUGAGAAUAAGGCUCCGACCUCUCGACCACGCCCCCAGGCGCACUGCCAAUCUCAAUCCCAUCUCCCACCAUCC ..((((((((.....)))))))).................(((.((((((((....(((..(((....)))....((((....))))....)))..))))))))....)))......... ( -29.50) >DroSec_CAF1 15054 108 + 1 AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGACGAGAAGAAGGCUCCGACCUCUGGACCACGCCCCCAGGCGCACUGCCAAUCUCAAUCCCAC------------ ..((((((((.....))))))))............(((......))).((((....(((((((.....))))...((((....))))....)))..))))........------------ ( -26.60) >DroSim_CAF1 15175 108 + 1 AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGAAGGCUCCGACCUCUGGACCACGCCCCCAGGCGCACUGCCAAUCUCAAUCCCAU------------ ..((((((((.....)))))))).................(((.((((.(((.(..(....)..).))).)))).((((....))))..........)))........------------ ( -28.20) >DroEre_CAF1 14692 108 + 1 AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGCAGAAGGCUCCGACCUCCGGACCACGCCCCCAGGCGUACUGCCAAUCUCAAUCCCGU------------ ..((((((((.....)))))))).................(((.((((((((.....))).)))))...((...(((((....)))))....))...)))........------------ ( -32.40) >DroYak_CAF1 14444 111 + 1 AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGGAGGCUCCGACCUCUGGACCACGCCCCCAGGCGCACUGCCAAUCUCAAUCCCGCCUC--------- ..((((((((.....))))))))...................((((..((((.(((((......)))))((....((((....)))).....))..))))....))))...--------- ( -32.40) >consensus AAUUUCGUGUAAAUAACACGAAAAUAACAAAAACAUCCUCGAGCGGUCGAGAAGAAGGCUCCGACCUCUGGACCACGCCCCCAGGCGCACUGCCAAUCUCAAUCCCAU____________ ..((((((((.....))))))))..............((((......)))).....(((((((.....))))...((((....))))....))).......................... (-25.02 = -24.90 + -0.12)

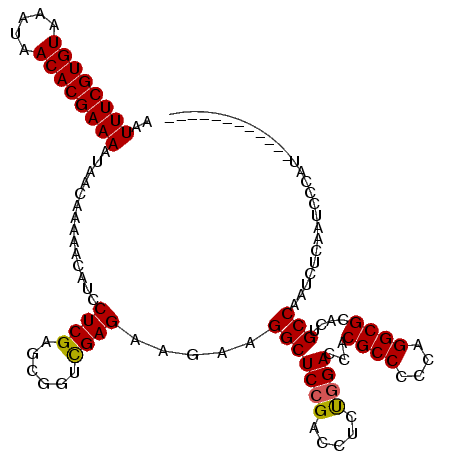
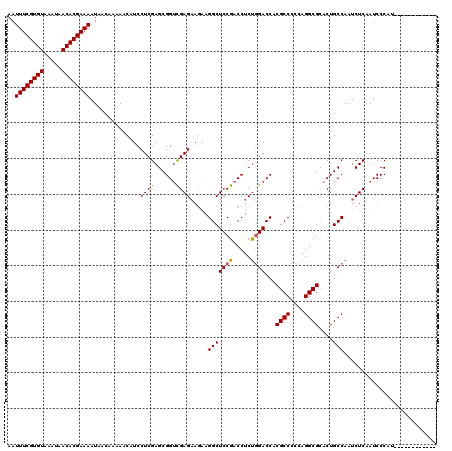
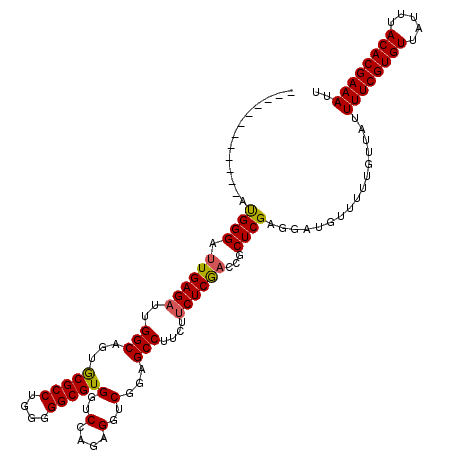
| Location | 2,365,546 – 2,365,666 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2365546 120 - 20766785 GGAUGGUGGGAGAUGGGAUUGAGAUUGGCAGUGCGCCUGGGGGCGUGGUCGAGAGGUCGGAGCCUUAUUCUCAACCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU .....(((..(((((((.((((((..(((...(((((....)))))(..(....)..)...)))....))))))))((.((((((((.......)))))))).))))))).)))...... ( -39.50) >DroSec_CAF1 15054 108 - 1 ------------GUGGGAUUGAGAUUGGCAGUGCGCCUGGGGGCGUGGUCCAGAGGUCGGAGCCUUCUUCUCGUCCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU ------------(((((((.((((.(((.(.((((((....)))))).))))(((((....)))))..))))))))((.((((((((.......)))))))).))......)))...... ( -35.50) >DroSim_CAF1 15175 108 - 1 ------------AUGGGAUUGAGAUUGGCAGUGCGCCUGGGGGCGUGGUCCAGAGGUCGGAGCCUUCUUCUCGACCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU ------------......(((((.((((.(.((((((....)))))).))))).((((((((.....))).))))).)))))...............((((((((.....)))))))).. ( -35.50) >DroEre_CAF1 14692 108 - 1 ------------ACGGGAUUGAGAUUGGCAGUACGCCUGGGGGCGUGGUCCGGAGGUCGGAGCCUUCUGCUCGACCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU ------------(((...(((((.((((.(.((((((....)))))).))))).(((((.(((.....)))))))).)))))...))).........((((((((.....)))))))).. ( -39.30) >DroYak_CAF1 14444 111 - 1 ---------GAGGCGGGAUUGAGAUUGGCAGUGCGCCUGGGGGCGUGGUCCAGAGGUCGGAGCCUCCUUCUCGACCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU ---------(((.(((..((((((.(((.(.((((((....)))))).))))(((((....)))))..)))))))))))).................((((((((.....)))))))).. ( -40.40) >consensus ____________AUGGGAUUGAGAUUGGCAGUGCGCCUGGGGGCGUGGUCCAGAGGUCGGAGCCUUCUUCUCGACCGCUCGAGGAUGUUUUUGUUAUUUUCGUGUUAUUUACACGAAAUU .............((((.((((((..(((...(((((....)))))(..(....)..)...)))....))))))...))))................((((((((.....)))))))).. (-33.30 = -33.14 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:18 2006