
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,859,422 – 12,859,544 |
| Length | 122 |
| Max. P | 0.938188 |

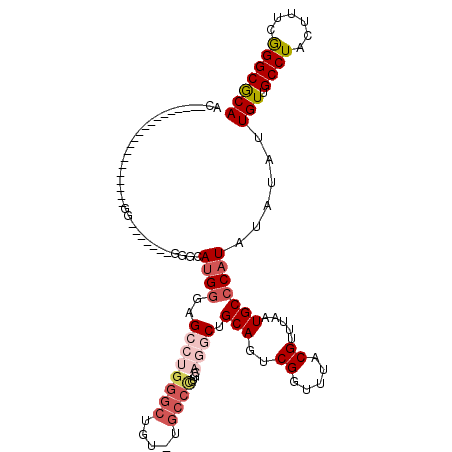
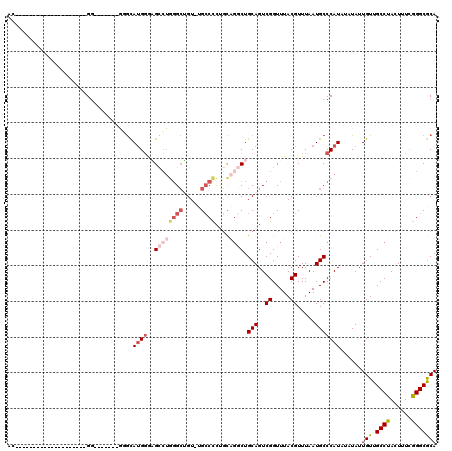
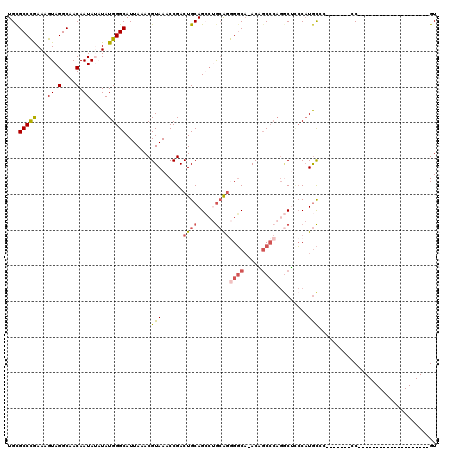
| Location | 12,859,422 – 12,859,514 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -16.30 |
| Energy contribution | -17.83 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
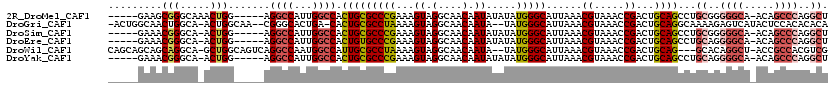
>2R_DroMel_CAF1 12859422 92 + 20766785 AC--------------------GG-------GUGCAUGGGAGCCUGGGCUGU-UGCCCCCGCAGGCUGCAGUCGGUUUACGUUUAAUGCCCAUAUAUAUUGUUGCCUACUUUCGGGCGCA ..--------------------((-------(((.(((..((((..((((((-.(((......))).))))))))))..)))....)))))........(((.((((......))))))) ( -30.90) >DroGri_CAF1 244230 111 + 1 ACACACACACAUAUAUAUACAUGU-------GUAUAUGUGUGUGUGUGGAGUAUGACUCUUUUGCCUGCAGUCGGUUUACGUUUAAUGCCCAUA--UAUUGUUGCCUACUUUUAGGCGCA .((((((((((((((((((....)-------)))))))))))))))))(((.....)))....(((((.(((.(((..(((.............--...))).))).)))..)))))... ( -40.39) >DroSim_CAF1 214786 92 + 1 AC--------------------GG-------GGGCAUGGGAGCCUGGGCUGU-UGCCCCCGCAGGCUGCAGUCGGUUUACGUUUAAUGCCCAUAUAUAUUGUUGCCUACUUUCGGGCGCA ..--------------------((-------(((((..(.(((....))).)-)))))))((.(((......((.....))......))).............((((......)))))). ( -33.90) >DroEre_CAF1 206963 92 + 1 AC--------------------GG-------GGGCAUGGCAGCCUGGGCUGU-UGCCCCUGCAGGCUGCAGUCGGUUUACGUUUAAUGCCCAUAUAUAUUGUUGCCUACUUUCGGGCACA ..--------------------..-------((((((.(((((((((((...-.))))....)))))))...((.....))....))))))...........(((((......))))).. ( -37.10) >DroWil_CAF1 311605 93 + 1 AC--------------------GGGUUACGACCGCA-GGACGACGUGGCGGU-AGCCUGUGC---CUGCAGUCGGUUUACGUUUAAUGCCCAUA--UAUUGUUGCCUACUUUUAGGCGCA ..--------------------((((..((((.(((-((.(.(((.(((...-.))))))))---)))).)))).............))))...--...(((.(((((....)))))))) ( -33.46) >DroYak_CAF1 212701 91 + 1 AC--------------------GG--------GGCAUGGAAGCCUGGGCUGU-UGCCCCUGCAGGCUGCAGUCGGUUUACGUUUAAUGCCCAUAUAUAUUGUUGCCUACUUUCGGGCGCA ..--------------------.(--------(((((..(((((..((((((-.(((......))).))))))))))).......))))))........(((.((((......))))))) ( -32.80) >consensus AC____________________GG_______GGGCAUGGGAGCCUGGGCUGU_UGCCCCUGCAGGCUGCAGUCGGUUUACGUUUAAUGCCCAUAUAUAUUGUUGCCUACUUUCGGGCGCA ...................................((((..((((((((.....))))....)))).(((..((.....)).....)))))))......(((.((((......))))))) (-16.30 = -17.83 + 1.53)

| Location | 12,859,422 – 12,859,514 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12859422 92 - 20766785 UGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCGGGGGCA-ACAGCCCAGGCUCCCAUGCAC-------CC--------------------GU (((((((....)..)))..........(((((........(((.......)))((((((.((.(...-.)..)))))))))))))))).-------..--------------------.. ( -28.50) >DroGri_CAF1 244230 111 - 1 UGCGCCUAAAAGUAGGCAACAAUA--UAUGGGCAUUAAACGUAAACCGACUGCAGGCAAAAGAGUCAUACUCCACACACACACAUAUAC-------ACAUGUAUAUAUAUGUGUGUGUGU ((((((((.(.((.(....).)).--).))))).......(((.......)))..)))...(((.....))).(((((((((((((((.-------.........))))))))))))))) ( -32.50) >DroSim_CAF1 214786 92 - 1 UGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCGGGGGCA-ACAGCCCAGGCUCCCAUGCCC-------CC--------------------GU ((((((((.(.((((....)..))).).)))))......((.....))...)))....(((((((((-..(((....)))....)))))-------))--------------------)) ( -32.70) >DroEre_CAF1 206963 92 - 1 UGUGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA-ACAGCCCAGGCUGCCAUGCCC-------CC--------------------GU (((((((....)..))).)))........((((((....((.....))...(((((((....((((.-...))))))))))).))))))-------..--------------------.. ( -34.10) >DroWil_CAF1 311605 93 - 1 UGCGCCUAAAAGUAGGCAACAAUA--UAUGGGCAUUAAACGUAAACCGACUGCAG---GCACAGGCU-ACCGCCACGUCGUCC-UGCGGUCGUAACCC--------------------GU ...(((((....))))).......--.(((((..(((....)))..(((((((((---((((.(((.-...)))..)).).))-))))))))...)))--------------------)) ( -32.40) >DroYak_CAF1 212701 91 - 1 UGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA-ACAGCCCAGGCUUCCAUGCC--------CC--------------------GU (((((((....)..)))............((((......((.....))......)))))))((((((-..(((....)))....))))--------))--------------------.. ( -28.40) >consensus UGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA_ACAGCCCAGGCUCCCAUGCCC_______CC____________________GU ...(((((...((.(....).)).....))))).......(((......((((.....))))((((.....)))).........)))................................. (-15.56 = -15.95 + 0.39)
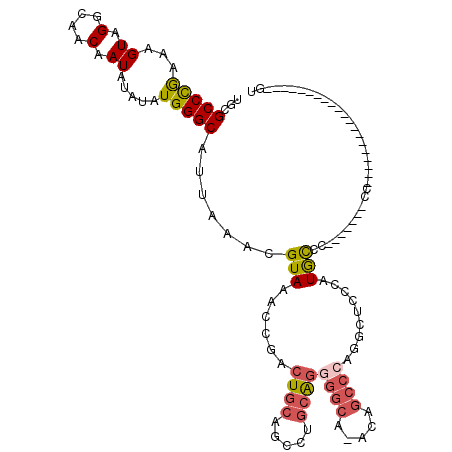
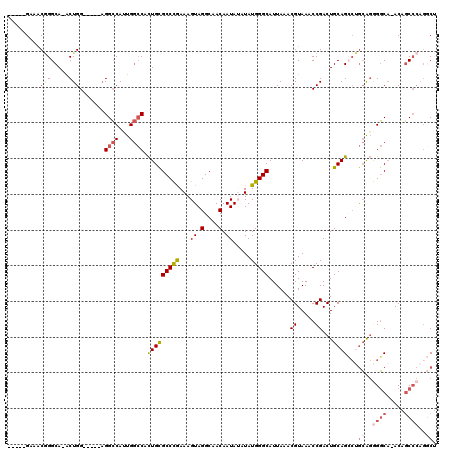
| Location | 12,859,435 – 12,859,544 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
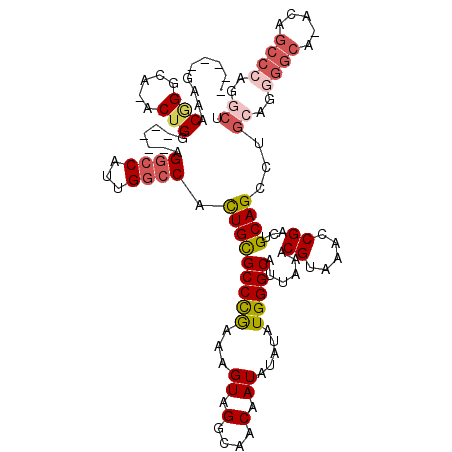
>2R_DroMel_CAF1 12859435 109 - 20766785 -----GAAGCGGGCAAACUGG-----AGGCCAUUGGCCACUGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCGGGGGCA-ACAGCCCAGGCU -----...((((((.......-----.((((...))))..((((((((.(.((((....)..))).).)))))......((.....))...)))))))))..((((.-...))))..... ( -37.70) >DroGri_CAF1 244263 113 - 1 -ACUGGCAACUGGCA-ACUGGCAA--CGGGCACUGA-CACUGCGCCUAAAAGUAGGCAACAAUA--UAUGGGCAUUAAACGUAAACCGACUGCAGGCAAAAGAGUCAUACUCCACACACA -..(((....((((.-.(((....--)))...((..-..(((((((((....))))).......--..(((..............)))...)))).....)).))))....)))...... ( -25.14) >DroSim_CAF1 214799 108 - 1 -----GAAACGGGCA-ACUGG-----AGGCCAUUGGCCACUGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCGGGGGCA-ACAGCCCAGGCU -----....(((...-.))).-----.((((...))))..((((((((.(.((((....)..))).).)))))......((.....))...)))(((((.((.(...-.)..))))))). ( -35.50) >DroEre_CAF1 206976 108 - 1 -----GAAACGGGCA-ACUGG-----AGGCCAUUGGCCACUGUGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA-ACAGCCCAGGCU -----....((((((-...((-----.((((...)))).)).))))))..(((.(....)........(((((................((((.....)))).(...-.).))))).))) ( -37.30) >DroWil_CAF1 311624 113 - 1 CAGCAGCAGCAGGCA-GCUGGCAGUCAGGCCAAUGGCCAUUGCGCCUAAAAGUAGGCAACAAUA--UAUGGGCAUUAAACGUAAACCGACUGCAG---GCACAGGCU-ACCGCCACGUCG ..((....)).(((.-(((.((((((.((((...))))..((((((((....)))))..((...--..)).))).............)))))).)---))...(((.-...)))..))). ( -39.30) >DroYak_CAF1 212713 108 - 1 -----GAAACGGGCA-ACUGG-----AGGCCAUUGGCCACUGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA-ACAGCCCAGGCU -----....(((...-.))).-----.((((...)))).(((((((((.(.((((....)..))).).)))))......((.....))...))))...((..((((.-...))))..)). ( -35.70) >consensus _____GAAACGGGCA_ACUGG_____AGGCCAUUGGCCACUGCGCCCGAAAGUAGGCAACAAUAUAUAUGGGCAUUAAACGUAAACCGACUGCAGCCUGCAGGGGCA_ACAGCCCAGGCU .........(((.....))).......((((...)))).(((((((((...((.(....).)).....)))))......((.....))...))))...((..((((.....))))..)). (-22.76 = -23.77 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:24 2006