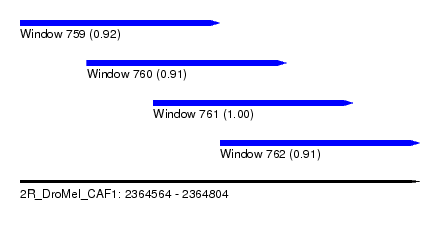
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,364,564 – 2,364,804 |
| Length | 240 |
| Max. P | 0.998781 |

| Location | 2,364,564 – 2,364,684 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.15 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -28.04 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364564 120 + 20766785 GAACCAGUGGACCCAGAACCCACUUGUUGGCCACGGUUCACCCGGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCA .((((((((......(....)..(((((((...(((..((...(((.(((((.((......)).))))))))...))..)))..)))))))...)))))))).................. ( -34.80) >DroSec_CAF1 14107 112 + 1 UC--------ACCCGAAACCCACUUGUUGGACACGGUUCACCCAGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCACCAACGACGCUGGUUGAUGUUUAACAAUUUGCA ..--------.............((((((((((....(((.(((((.(((((.((......)).))))).....(((((.......)))))....))))).)))))))))))))...... ( -31.80) >DroSim_CAF1 14202 112 + 1 UC--------ACCCGAAACCCACUUGUUGGACACGGUUCACCCAGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCGACAACGACGCUGGUUGAUGUUUAACAAUUUGCA ..--------.............((((((((((....(((.(((((.(((((.((......)).))))).....((((......)))).......))))).)))))))))))))...... ( -30.70) >DroEre_CAF1 13771 112 + 1 UC--------ACCCGGAACCCACUUGUUGGACACGGUUCACCCGGCAGCGAGCUCUUAAUUGAUUUCGCGCUAAUUGGUGCGACCCACCAGCGACGCUGGUUGAUGUUUAACAAUUUGCA ..--------.............((((((((((.((((((((.(((.(((((.((......)).))))))))....)))).)))).((((((...))))))...))))))))))...... ( -35.80) >DroYak_CAF1 13510 112 + 1 UU--------ACCCAGAACCCACUUGUUGGUCACGGUUCAUCCGGCAGCGAACUCUUAAUUGAUUUCGUGCUAAUUGGUCCGACCCAACAACUACGCUGGUUGAUGUUUAACAAUUUGCA ((--------(.((((.......(((((((...(((..((...((((.((((.((......)).))))))))...))..)))..))))))).....)))).)))................ ( -29.30) >consensus UC________ACCCGGAACCCACUUGUUGGACACGGUUCACCCGGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCA .......................((((((((((....(((.(((((.(((((.((......)).)))))......(((......)))........))))).)))))))))))))...... (-28.04 = -27.72 + -0.32)

| Location | 2,364,604 – 2,364,724 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364604 120 + 20766785 CCCGGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCCC .(((((.(((((.((......)).))))).....((((......)))).......))))).(((((((((..............))))))))).....((.(((((....)))))..)). ( -27.84) >DroSec_CAF1 14139 120 + 1 CCCAGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCACCAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGC .(((((.(((((.((......)).))))).....(((((.......)))))....))))).(((((((((..............)))))))))....(((.(((((....))))).))). ( -30.04) >DroSim_CAF1 14234 120 + 1 CCCAGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCGACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGC .(((((.(((((.((......)).)))))))(((((((((......)))...(((((((..(((((((((..............)))))))))....))))))))))))))))....... ( -30.74) >DroEre_CAF1 13803 120 + 1 CCCGGCAGCGAGCUCUUAAUUGAUUUCGCGCUAAUUGGUGCGACCCACCAGCGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGC .......(((((.(((((((((...(((((((....))))))).........(((((((..(((((((((..............)))))))))....))))))))))))).))).))))) ( -33.84) >DroYak_CAF1 13542 120 + 1 UCCGGCAGCGAACUCUUAAUUGAUUUCGUGCUAAUUGGUCCGACCCAACAACUACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCCACGGUGUCCAAUUAUGGAUUUCGC (((((((.((((.((......)).)))))))(((((((.(((..(((.(......).))).(((((((((..............)))))))))....)))...)))))))))))...... ( -26.34) >consensus CCCGGCAGCGAACUCUUAAUUGAUUUCGCGCUAAUUGGUCCGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGC .(((((.(((((.((......)).)))))......(((......)))........))))).(((((((((..............)))))))))....(((.(((((....))))).))). (-27.06 = -26.54 + -0.52)

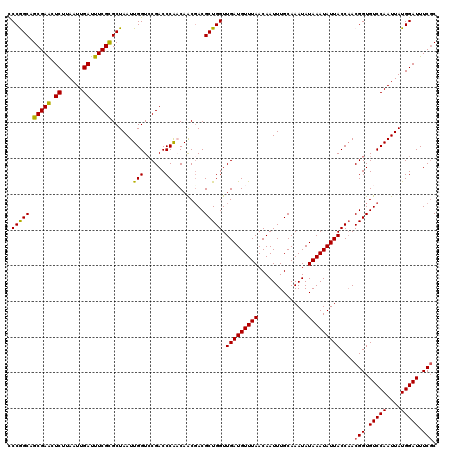
| Location | 2,364,644 – 2,364,764 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364644 120 + 20766785 CGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCCCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGG ...((((.....(((((((..(((((((((..............)))))))))....)))))))......(((((.....(((((((....)))))))...)))))..........)))) ( -28.64) >DroSec_CAF1 14179 120 + 1 CGACCCACCAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGG ...((((.....(((((((..(((((((((..............)))))))))....)))))))......(((((...(.(((((((....))))))))..)))))..........)))) ( -28.64) >DroSim_CAF1 14274 120 + 1 CGACCCGACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGG ...(((((((....((.((((.((((((((..............)))))))))))).)).))))......(((((...(.(((((((....))))))))..)))))...........))) ( -28.74) >DroEre_CAF1 13843 120 + 1 CGACCCACCAGCGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGG ...((((...((((((.((((.((((((((..............)))))))))))).))..(((((....))))).))))(((((((....)))))))..................)))) ( -31.44) >DroYak_CAF1 13582 120 + 1 CGACCCAACAACUACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCCACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGA ....(((........(((((.(((((((((..............))))))))).))).((.(((((....))))).))))(((((((....)))))))..................))). ( -25.44) >consensus CGACCCAACAACGACGCUGGUUGAUGUUUAACAAUUUGCAAAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGG ...((((.....(((((((..(((((((((..............)))))))))....)))))))......(((((.....(((((((....)))))))...)))))..........)))) (-26.48 = -26.72 + 0.24)

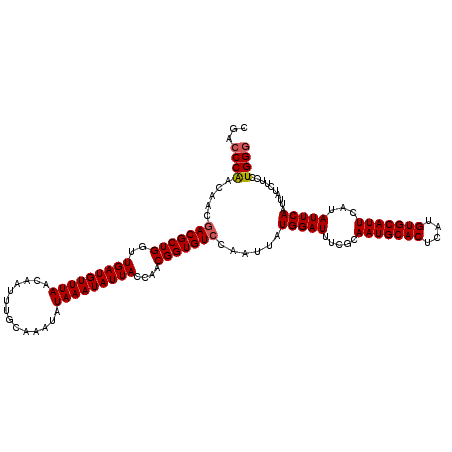
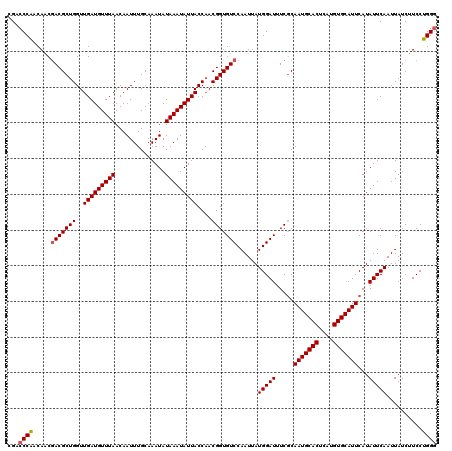
| Location | 2,364,684 – 2,364,804 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364684 120 + 20766785 AAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCCCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGGCGGGCUUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .............(((..((.(((((....)))))...))(((((((....)))))))..................))).(.(((((((..(((.......)))....))))))).)... ( -28.10) >DroSec_CAF1 14219 120 + 1 AAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGGCGGGCAUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .................(((((((((....))))).(((((((((((....))))))........................(((((.......))))).........))))))))).... ( -29.30) >DroSim_CAF1 14314 120 + 1 AAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGGCGGGCAUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .................(((((((((....))))).(((((((((((....))))))........................(((((.......))))).........))))))))).... ( -29.30) >DroEre_CAF1 13883 120 + 1 AAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGGCGGGCAUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .................(((((((((....))))).(((((((((((....))))))........................(((((.......))))).........))))))))).... ( -29.30) >DroYak_CAF1 13622 120 + 1 AAUAUAAAUAUUACCCACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGACGGGCUUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .................(((((((((....))))).(((((((((((....)))))).......((((((.(..(((....)))...).))))))............))))))))).... ( -28.70) >consensus AAUAUAAAUAUUACCAACGGUGUCCAAUUAUGGAUUUCGCAAUGCACUCAUGUGCAUUCAUAUUCAAUUAUCUUCCUGGGCGGGCAUGUUAAUUGCUCUUAAAUCUGUGCGAGCCGGAAA .................(((((((((....))))).(((((((((((....)))))).......((((((.(..(((....)))...).))))))............))))))))).... (-27.34 = -27.54 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:14 2006