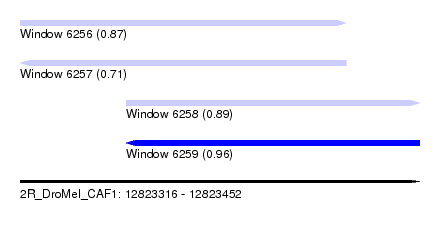
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,823,316 – 12,823,452 |
| Length | 136 |
| Max. P | 0.961841 |

| Location | 12,823,316 – 12,823,427 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12823316 111 + 20766785 ---AUUCAUGCAUUUUAGU-UUCAAAUUACUUUUAUUAUUCGAAUAAAA-AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGUGCAGAUUUCGGCGCCACCUCAGCACAUCU ---.....((((((((((.-..........(((((((.....)))))))-..((((.....))))......)))))))....(((((((.(....).)))))))....)))..... ( -21.20) >DroSec_CAF1 167302 93 + 1 ---UUUCAUGCAUUUUAGU-UUCAA------------------AUAAAA-AAUGCCAAAUUGGUAAUUUCUUAAAAAUUUAUGUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCU ---.....(((........-.....------------------......-..((((.....)))).................(((((((.(....).)))))))....)))..... ( -20.40) >DroSim_CAF1 172615 93 + 1 ---UUUCAUGCAUUUUAGU-UUCAA------------------AUAAAA-AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGCGCAGAAUUCGGCGCCACCUCAGCACAUCU ---.....(((........-.....------------------......-..((((.....)))).................(((((((.(....).)))))))....)))..... ( -20.40) >DroEre_CAF1 170195 97 + 1 CAGUAUCAC-CAUUUUAUUAUUCAA------------------AUAAAACAAUGCCGGAUUGGUAAUUUCUUUGAAAAUUAUUUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCU .........-..(((((((.....)------------------))))))...(((.((...((((((((......))))))))((((((.(....).))))))))...)))..... ( -17.10) >DroYak_CAF1 175162 97 + 1 CAGCAUUAU-CCUUUUAUUAUUCAA------------------AUAAAGGAAUGCCAAACUGGUAAUUUCUUUGAACAUUAUUCGGCGCAGAUUUCGGCGCCACCUCAGCACAUCU ..((.....-...............------------------.((((((((((((.....))).)))))))))..........(((((.(....).)))))......))...... ( -22.70) >consensus ___UUUCAUGCAUUUUAGU_UUCAA__________________AUAAAA_AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCU ....................................................((((.....)))).................(((((((.(....).)))))))............ (-14.52 = -14.96 + 0.44)

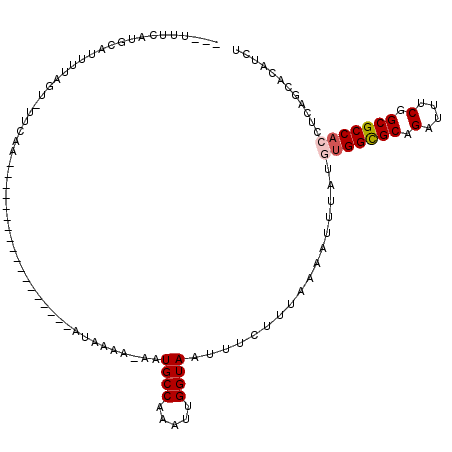
| Location | 12,823,316 – 12,823,427 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12823316 111 - 20766785 AGAUGUGCUGAGGUGGCGCCGAAAUCUGCACCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU-UUUUAUUCGAAUAAUAAAAGUAAUUUGAA-ACUAAAAUGCAUGAAU--- ............((((.((.(....).)).)))).((((((...((....))...))))))(((((-((...(((((((.((....)).)))))))-...)))))))......--- ( -20.50) >DroSec_CAF1 167302 93 - 1 AGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCACAUAAAUUUUUAAGAAAUUACCAAUUUGGCAUU-UUUUAU------------------UUGAA-ACUAAAAUGCAUGAAA--- ............(((((((.(....).))))))).((((((..(((....)))..))))))(((((-((....------------------.....-...)))))))......--- ( -22.30) >DroSim_CAF1 172615 93 - 1 AGAUGUGCUGAGGUGGCGCCGAAUUCUGCGCCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU-UUUUAU------------------UUGAA-ACUAAAAUGCAUGAAA--- ....((((..(((((((((.(....).))))))).....(((((((((((...((.....))...)-)))..)------------------)))))-))).....))))....--- ( -21.60) >DroEre_CAF1 170195 97 - 1 AGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCAAAUAAUUUUCAAAGAAAUUACCAAUCCGGCAUUGUUUUAU------------------UUGAAUAAUAAAAUG-GUGAUACUG ......((((.((((((((.(....).)))))...(((((((....)))))))...)))))))((..((((((------------------(.....)))))))..-))....... ( -24.10) >DroYak_CAF1 175162 97 - 1 AGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCGAAUAAUGUUCAAAGAAAUUACCAGUUUGGCAUUCCUUUAU------------------UUGAAUAAUAAAAGG-AUAAUGCUG ......((((....(((((.(....).)))))((((...))))...........))))..(((((((((((((------------------(.....))).)))))-)..))))). ( -23.70) >consensus AGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU_UUUUAU__________________UUGAA_ACUAAAAUGCAUGAAA___ ....(((((((((((((((.(....).)))))))..........((....)).....))))))))................................................... (-16.28 = -16.76 + 0.48)


| Location | 12,823,352 – 12,823,452 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12823352 100 + 20766785 CGAAUAAAA-AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGUGCAGAUUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU .((......-..((((.....)))).................(((((((.(....).))))))).))(((..((((..(((....)))..))))...))). ( -20.00) >DroSec_CAF1 167323 97 + 1 ---AUAAAA-AAUGCCAAAUUGGUAAUUUCUUAAAAAUUUAUGUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU ---......-..((((.....)))).................(((((((.(....).)))))))...(((..((((..(((....)))..))))...))). ( -21.70) >DroSim_CAF1 172636 97 + 1 ---AUAAAA-AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGCGCAGAAUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU ---......-..((((.....)))).................(((((((.(....).)))))))...(((..((((..(((....)))..))))...))). ( -21.70) >DroEre_CAF1 170219 98 + 1 ---AUAAAACAAUGCCGGAUUGGUAAUUUCUUUGAAAAUUAUUUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU ---.........(((.((...((((((((......))))))))((((((.(....).))))))))...)))........((((((((....))))).))). ( -19.00) >DroYak_CAF1 175186 98 + 1 ---AUAAAGGAAUGCCAAACUGGUAAUUUCUUUGAACAUUAUUCGGCGCAGAUUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU ---.((((((((((((.....))).)))))))))..........(((((.(....).))))).....(((..((((..(((....)))..))))...))). ( -24.50) >consensus ___AUAAAA_AAUGCCAAAUUGGUAAUUUCUUUAAAAUUUAUGUGGCGCAGAUUUCGGCGCCACCUCAGCACAUCUCUCAGCAAAUUGUUAGAUUUCGCUU ............((((.....)))).................(((((((.(....).)))))))...(((..((((..(((....)))..))))...))). (-18.92 = -19.36 + 0.44)

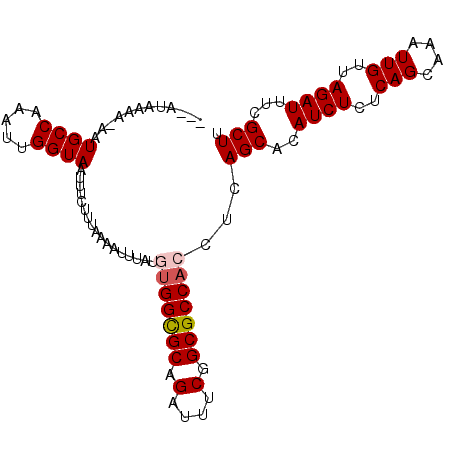

| Location | 12,823,352 – 12,823,452 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12823352 100 - 20766785 AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAAUCUGCACCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU-UUUUAUUCG .((((((.........))))))..((((.((((..((((((.((.(....).)).))))...((((((...)))))).....))..)))).-))))..... ( -21.10) >DroSec_CAF1 167323 97 - 1 AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCACAUAAAUUUUUAAGAAAUUACCAAUUUGGCAUU-UUUUAU--- .((((((.........))))))(((((((((......(((((((.(....).))))))).((((((..(((....)))..)))))))))))-))))..--- ( -27.90) >DroSim_CAF1 172636 97 - 1 AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAUUCUGCGCCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU-UUUUAU--- .((((((.........))))))..((((.((((..(((((((((.(....).)))))))...((((((...)))))).....))..)))).-))))..--- ( -26.50) >DroEre_CAF1 170219 98 - 1 AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCAAAUAAUUUUCAAAGAAAUUACCAAUCCGGCAUUGUUUUAU--- .((((((.........))))))..(((((((((((.((((((((.(....).)))))...(((((((....)))))))...)))))))).))))))..--- ( -24.80) >DroYak_CAF1 175186 98 - 1 AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCGAAUAAUGUUCAAAGAAAUUACCAGUUUGGCAUUCCUUUAU--- .((((((.........)))))).((((..((((..(((((((((.(....).)))))((((...)))).........)))...)..))))..))))..--- ( -25.90) >consensus AAGCGAAAUCUAACAAUUUGCUGAGAGAUGUGCUGAGGUGGCGCCGAAAUCUGCGCCACAUAAAUUUUAAAGAAAUUACCAAUUUGGCAUU_UUUUAU___ .((((((.........)))))).((((..(((((((((((((((.(....).)))))))..........((....)).....))))))))..))))..... (-20.80 = -21.12 + 0.32)

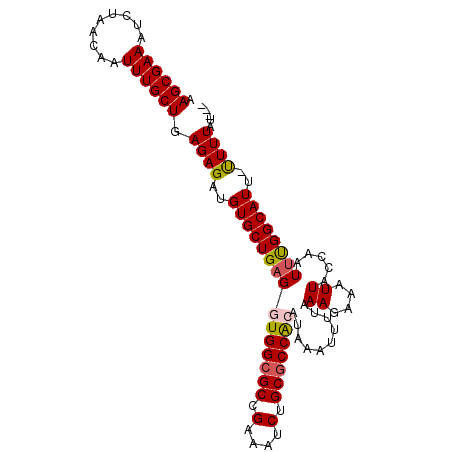
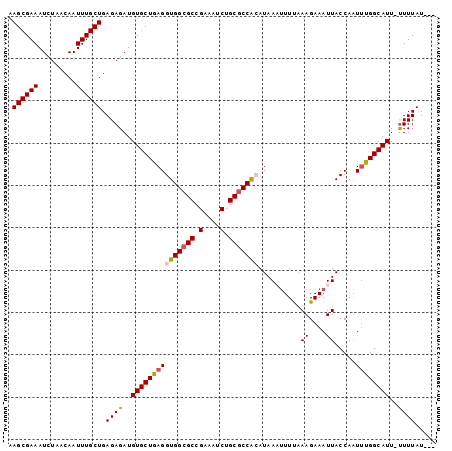
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:11 2006