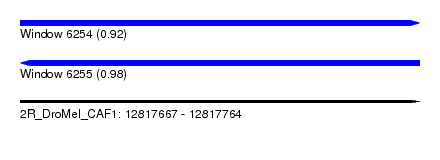
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,817,667 – 12,817,764 |
| Length | 97 |
| Max. P | 0.975268 |

| Location | 12,817,667 – 12,817,764 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12817667 97 + 20766785 GCAGACAACAGACAACAGACU-ACAGACAGCCAG-------ACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAG---CAGCUAGCCAUGGCUGUCA .....................-...((((((((.-------......(((........)))...((((.((((....)))).))))---..........)))))))). ( -27.90) >DroSec_CAF1 161630 91 + 1 GCAGACAGC------AAGACU-ACAGACAGCCAG-------ACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAG---CAGCUAGCCAAGGCUGUCA ((.....))------......-...(((((((..-------......(((........)))...((((.((((....)))).))))---...........))))))). ( -28.10) >DroSim_CAF1 166882 98 + 1 GCAGACAGC------CAGACU-ACAGACAGCCAGACUACAGACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAG---CAGCUAGCCAAUGCUGUCA ...((((((------..(.((-(......((((((.........))))))........((....((((.((((....)))).))))---..))))).)...)))))). ( -26.10) >DroEre_CAF1 164372 98 + 1 ACAGACAACAGCCAACAGACUGGCAGACUGGCAG-------ACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAG---CAGCUAGCCAAAGCUGUCA ...((((.(.((((......)))).(.(((((..-------......(((........)))...((((.((((....)))).))))---..))))).)...).)))). ( -30.40) >DroYak_CAF1 169225 79 + 1 AC----------------------GGACUGCCAG-------ACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAGCAGCAGCUAGCCAAAGCUGUCA ..----------------------((....)).(-------(((((((((..((.((.((....((((.((((....)))).))))..)))).))))))))..)))). ( -22.60) >consensus GCAGACAAC______CAGACU_ACAGACAGCCAG_______ACAUUUGGCCAUAUCUCGCCUUCCUGCUCUUGACCACAAGUGCAG___CAGCUAGCCAAAGCUGUCA .........................((((((................(((........)))...((((.((((....)))).))))...............)))))). (-19.16 = -19.76 + 0.60)

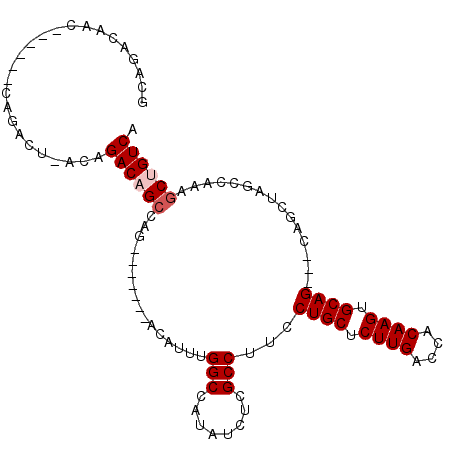
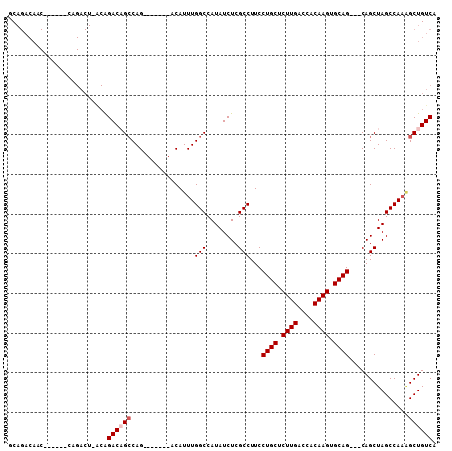
| Location | 12,817,667 – 12,817,764 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12817667 97 - 20766785 UGACAGCCAUGGCUAGCUG---CUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGU-------CUGGCUGUCUGU-AGUCUGUUGUCUGUUGUCUGC .((((((.(..((..((((---((((.((((....)))).)))(((.(((.((((((......))))-------)).))).)))))-)))..))..)..))))))... ( -37.30) >DroSec_CAF1 161630 91 - 1 UGACAGCCUUGGCUAGCUG---CUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGU-------CUGGCUGUCUGU-AGUCUU------GCUGUCUGC .((((((...(((((....---((((.((((....)))).))))((.(((.((((((......))))-------)).))).))..)-))))..------))))))... ( -37.20) >DroSim_CAF1 166882 98 - 1 UGACAGCAUUGGCUAGCUG---CUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGUCUGUAGUCUGGCUGUCUGU-AGUCUG------GCUGUCUGC .((((((...((((((((.---((((.((((....)))).))))...))).((((..(((((.((.......)).))))))))).)-))))..------))))))... ( -37.70) >DroEre_CAF1 164372 98 - 1 UGACAGCUUUGGCUAGCUG---CUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGU-------CUGCCAGUCUGCCAGUCUGUUGGCUGUUGUCUGU .((((((...(((((((.(---(((..((((....)))).(((((..(((.((((((......))))-------)))))..)))))))))..)))))))))))))... ( -40.60) >DroYak_CAF1 169225 79 - 1 UGACAGCUUUGGCUAGCUGCUGCUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGU-------CUGGCAGUCC----------------------GU ...(((((......)))))....(((.((((....)))).)))(((..((.((((((......))))-------)).))..)))----------------------.. ( -28.70) >consensus UGACAGCCUUGGCUAGCUG___CUGCACUUGUGGUCAAGAGCAGGAAGGCGAGAUAUGGCCAAAUGU_______CUGGCUGUCUGU_AGUCUG______GCUGUCUGC .(((((((((((((((((....((((.((((....)))).))))...)))......))))))).............)))))))......................... (-24.99 = -25.83 + 0.84)

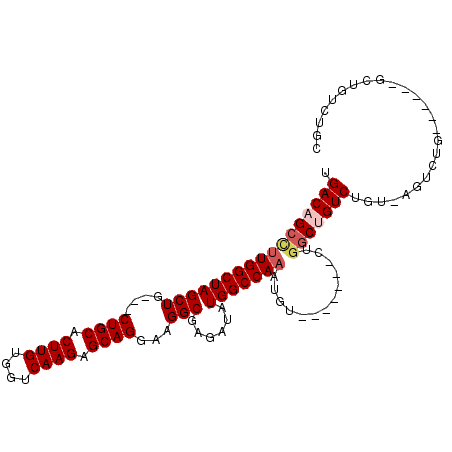
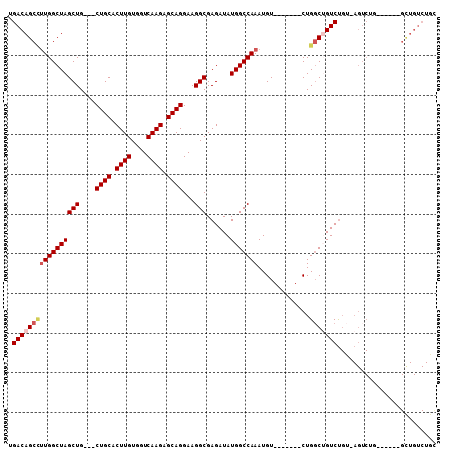
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:08 2006