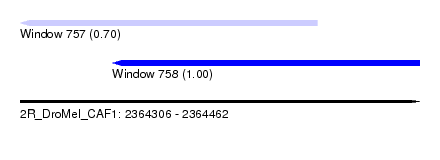
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,364,306 – 2,364,462 |
| Length | 156 |
| Max. P | 0.999386 |

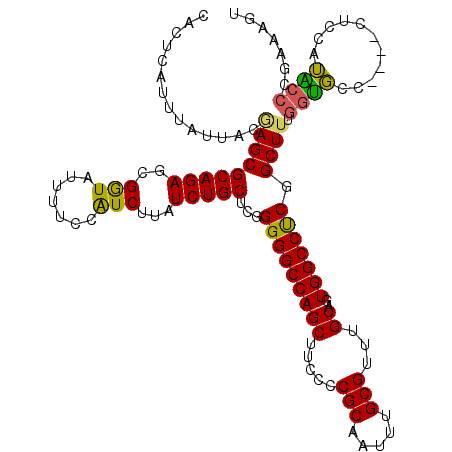
| Location | 2,364,306 – 2,364,422 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -29.20 |
| Energy contribution | -28.36 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364306 116 - 20766785 CACUCAUUUAUUACAAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCAGUGGCCUCGGCUUUGGUGCU----CUCCAUACCCGAAAGU .......(((((.((((((((((((((...............))))))(((((......)))))......)))))))).)))))..((((...(((....----..)))...)))).... ( -36.56) >DroSec_CAF1 13842 116 - 1 CACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCAGUGGCCUCGGCUUUGGUGCG----CUCCAUACCCGAAAGU ..............(((((((((((((...............)))))).(((((((((.....(((.....)))...))..)))))))........))))----)))......(....). ( -38.06) >DroSim_CAF1 13936 116 - 1 CACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCAGUGGCCUCGGCUUUGGUGCG----CUCCAUACCCGAAAGU ..............(((((((((((((...............)))))).(((((((((.....(((.....)))...))..)))))))........))))----)))......(....). ( -38.06) >DroEre_CAF1 13540 119 - 1 CACUCAUUUGUUACAAGCGCAGAGCGAUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCC-CGCAAUUUGCGUUUGCAUUGGCCCCAGCUCCCAGCUCCAAACUCCAGUCCCAAAAGU ......((((..((..(.(((((..(((.......)))...))))).).(((((((((....-(((.....)))...))..)))))))(((.....)))..........))..))))... ( -32.90) >DroYak_CAF1 13288 116 - 1 CACUCAUUUAUUACGAGCGCAGAGUGAUAUUUUCCGUCUUAUCUGCUCGGGGGCCAGCUUCCUCGCAAUACGCGUUUGCAUUGGCCUCGGCUUGGGUGUC----CUCCAUGCCCAAAAGU ..............(((((((((..(((.......)))...)))))...(((((((((.....(((.....)))...))..))))))).))))((((((.----....))))))...... ( -36.60) >consensus CACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCAGUGGCCUCGGCUUUGGUGCC____CUCCAUACCCGAAAGU ..............(((((((((..(((.......)))...)))))...(((((((((.....(((.....)))...))..))))))).)))).((((...........))))....... (-29.20 = -28.36 + -0.84)

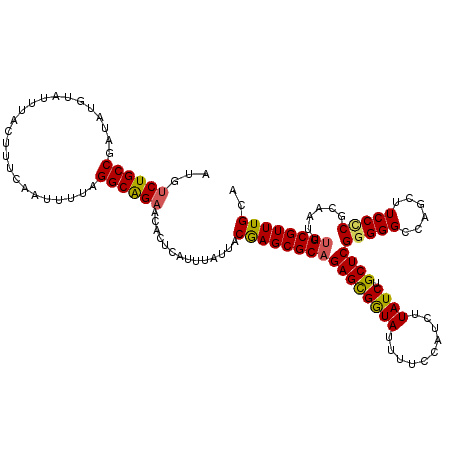
| Location | 2,364,342 – 2,364,462 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.12 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2364342 120 - 20766785 AUGUCUGCCGAUAUGUAUUUACUUUCAAUUUUAGGCAGUACACUCAUUUAUUACAAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCA .((((((((........................))))).)))...........((((((((((((((...............))))))(((((......)))))......)))))))).. ( -35.72) >DroSec_CAF1 13878 120 - 1 AUAUCUGCCGAUAUGUAUUUACUUUCAAUUUUAGGCAGAACACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCA ...((((((........................))))))..............((((((((((((((...............))))))(((((......)))))......)))))))).. ( -35.22) >DroSim_CAF1 13972 120 - 1 AUAUCUGCCGAUAUGUAUUUACUUUCAAUUUUAGGCAGAACACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCA ...((((((........................))))))..............((((((((((((((...............))))))(((((......)))))......)))))))).. ( -35.22) >DroEre_CAF1 13580 119 - 1 AUGUCUGCCGAUAUGUAUUUACUUUCAAUUUUAGGCGGAACACUCAUUUGUUACAAGCGCAGAGCGAUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCC-CGCAAUUUGCGUUUGCA .((((((((........................))))).)))...........((((((((((((((((..........)))).))))(((((....)))))-.......)))))))).. ( -32.06) >DroYak_CAF1 13324 120 - 1 AUGUCUGCCGAUACGUAUUUACUUUCAAUUUUAGGCAGAACACUCAUUUAUUACGAGCGCAGAGUGAUAUUUUCCGUCUUAUCUGCUCGGGGGCCAGCUUCCUCGCAAUACGCGUUUGCA .((((((((........................))))).)))...........(((((((.((((((((..........)))).))))(((((....))))).........))))))).. ( -30.36) >consensus AUGUCUGCCGAUAUGUAUUUACUUUCAAUUUUAGGCAGAACACUCAUUUAUUACGAGCGCAGAGCGGUAUUUUCCAUCUUAUCUGCUCGGGGGCCAGCUUCCCCGCAAUUUGCGUUUGCA ...((((((........................))))))..............((((((((((((((((..........)))).))))(((((......)))))......)))))))).. (-31.44 = -31.12 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:10 2006