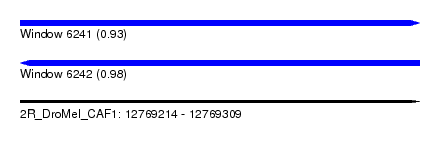
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,769,214 – 12,769,309 |
| Length | 95 |
| Max. P | 0.978904 |

| Location | 12,769,214 – 12,769,309 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
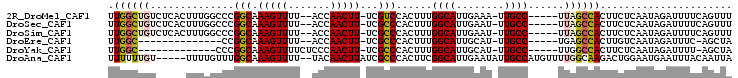
>2R_DroMel_CAF1 12769214 95 + 20766785 UUGGCUGUCUCACUUUGGCCCGGCAAAGUUUU--ACCAACUU-UCGUCCACUUUGGCAUUGAAA-UUGCC-----UUAGCCACUUCUCAAUAGAUUUUCAGUUU .((((((..............(((((((((..--...)))))-).)))......((((......-.))))-----.))))))...................... ( -17.90) >DroSec_CAF1 110849 95 + 1 UUGGCUGUCUCACUUUGGCCCGGCAAAGUUUU--ACCAACUU-UCGCCCACUUUGGCAUUGAAU-UUGCC-----UUAGCCACUUCUCAAUAGAUUUUCAGUUU .((((((..............(((((((((..--...)))))-).)))......((((......-.))))-----.))))))...................... ( -20.60) >DroSim_CAF1 118736 95 + 1 UUGGCUGUCUCACUUUGGCCCGGCAAAGUUUU--ACCAACUU-UCGCCCACUUUGGCAUUGAAU-UUGCC-----UUAGCCACUUCUCAAUAGAUUUUCAGUUU .((((((..............(((((((((..--...)))))-).)))......((((......-.))))-----.))))))...................... ( -20.60) >DroEre_CAF1 114231 80 + 1 UUGGC--------------CCGGCAAAGUUUU--ACCAACUU-UCGCCCACUUUGGCAUUGCAU-UUGCC-----UGAGCCACUUGUCAAUAGAUUUC-AGCUA .((((--------------.((((((((((..--...)))))-).)))......((((......-.))))-----.).))))................-..... ( -18.60) >DroYak_CAF1 118962 83 + 1 UUGGC-------------CCCGGCAAAGUUUUCUCCCAACUU-UCGCCCACUUUGGCAUUGCAU-UUGCC-----UUGGCCACUUCUCAAUAGAUUUU-AGCUA .((((-------------(..(((((((((.......)))))-).)))......((((......-.))))-----..)))))................-..... ( -21.00) >DroAna_CAF1 103077 97 + 1 UUUUUUGU-----UUUUGUUUGGCAAAGUUUU--UACAACUUAUCGCCCACUUCGGCAUUGAAUAUUGCCAUGUUUUGGCAAGACUGGAAUGAAUUUACAAUUA ...((..(-----((..(((((((((((((..--...)))))...(((......))).........)))).(((....)))))))..)))..)).......... ( -17.60) >consensus UUGGCUGU_____UUUGGCCCGGCAAAGUUUU__ACCAACUU_UCGCCCACUUUGGCAUUGAAU_UUGCC_____UUAGCCACUUCUCAAUAGAUUUUCAGUUA .((((((..............(((.(((((.......)))))...)))......((((........))))......))))))...................... (-13.97 = -14.17 + 0.20)
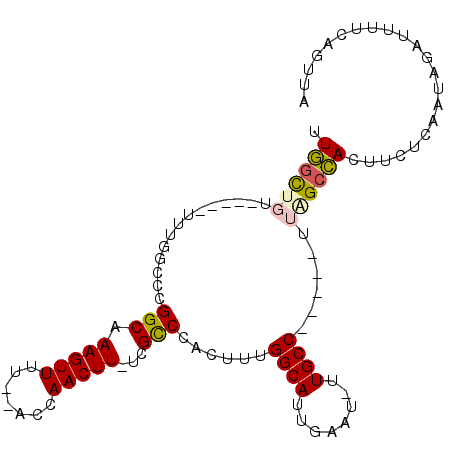

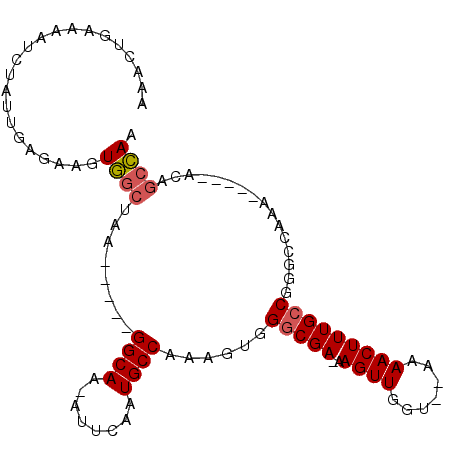
| Location | 12,769,214 – 12,769,309 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12769214 95 - 20766785 AAACUGAAAAUCUAUUGAGAAGUGGCUAA-----GGCAA-UUUCAAUGCCAAAGUGGACGA-AAGUUGGU--AAAACUUUGCCGGGCCAAAGUGAGACAGCCAA ...........(((((....)))))....-----(((..-(((((..(((......(((..-..)))(((--((....))))).))).....)))))..))).. ( -22.90) >DroSec_CAF1 110849 95 - 1 AAACUGAAAAUCUAUUGAGAAGUGGCUAA-----GGCAA-AUUCAAUGCCAAAGUGGGCGA-AAGUUGGU--AAAACUUUGCCGGGCCAAAGUGAGACAGCCAA ...(((....(((....)))..(((((..-----((((.-......))))......(((.(-(((((...--..))))))))).)))))........))).... ( -24.20) >DroSim_CAF1 118736 95 - 1 AAACUGAAAAUCUAUUGAGAAGUGGCUAA-----GGCAA-AUUCAAUGCCAAAGUGGGCGA-AAGUUGGU--AAAACUUUGCCGGGCCAAAGUGAGACAGCCAA ...(((....(((....)))..(((((..-----((((.-......))))......(((.(-(((((...--..))))))))).)))))........))).... ( -24.20) >DroEre_CAF1 114231 80 - 1 UAGCU-GAAAUCUAUUGACAAGUGGCUCA-----GGCAA-AUGCAAUGCCAAAGUGGGCGA-AAGUUGGU--AAAACUUUGCCGG--------------GCCAA .....-................((((((.-----((((.-......))))......(((.(-(((((...--..)))))))))))--------------)))). ( -24.90) >DroYak_CAF1 118962 83 - 1 UAGCU-AAAAUCUAUUGAGAAGUGGCCAA-----GGCAA-AUGCAAUGCCAAAGUGGGCGA-AAGUUGGGAGAAAACUUUGCCGGG-------------GCCAA .....-....(((....)))..(((((..-----(((..-......((((......))))(-(((((.......)))))))))..)-------------)))). ( -25.00) >DroAna_CAF1 103077 97 - 1 UAAUUGUAAAUUCAUUCCAGUCUUGCCAAAACAUGGCAAUAUUCAAUGCCGAAGUGGGCGAUAAGUUGUA--AAAACUUUGCCAAACAAAA-----ACAAAAAA ...((((.....((((..(((.((((((.....)))))).))).))))........(((((..((((...--..)))))))))..))))..-----........ ( -17.40) >consensus AAACUGAAAAUCUAUUGAGAAGUGGCUAA_____GGCAA_AUUCAAUGCCAAAGUGGGCGA_AAGUUGGU__AAAACUUUGCCGGGCCAAA_____ACAGCCAA ......................((((........((((........))))......(((((..((((.......)))))))))................)))). (-16.18 = -16.40 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:55 2006