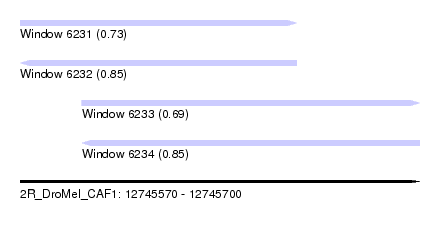
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,745,570 – 12,745,700 |
| Length | 130 |
| Max. P | 0.846312 |

| Location | 12,745,570 – 12,745,660 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -15.54 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12745570 90 + 20766785 ------UUUUUCACUGGCUCUUGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUUCCCGUAAUCCCAUUAAG ------.........(((.....)))...(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... ( -24.50) >DroSec_CAF1 92027 90 + 1 ------UUUUUCACUGGCUCUUGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAG ------.........(((.....)))...(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... ( -24.50) >DroSim_CAF1 99292 90 + 1 ------UUUUUCACUGGCUCUUGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAG ------.........(((.....)))...(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... ( -24.50) >DroEre_CAF1 93241 91 + 1 -----UUUUUUCGCUGACUCUCGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCCAUGGAGAUUAAUCCCCGUAAUCCCAUUAAG -----.......((((.....))))....(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... ( -27.40) >DroYak_CAF1 99898 96 + 1 UUCUUUUUUUGCACUGCCUCUUGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAG ..........(.((.(((....))).)))(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... ( -27.60) >DroAna_CAF1 86286 87 + 1 UUUUUUUUUUUUGUGUUCUGCUGUCAGUCACUGAAGAUCGGUUUAGGGGUUUUCCUUCCGUCGUCCCAGGCGAUUAAUCGAGGAAAU--------- ......(((((.(((..(((....))).))).)))))((((((..((((......))))(((((.....))))).))))))......--------- ( -16.70) >consensus ______UUUUUCACUGGCUCUUGGCUGUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAG ...............(((........)))(((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).... (-15.54 = -17.52 + 1.97)

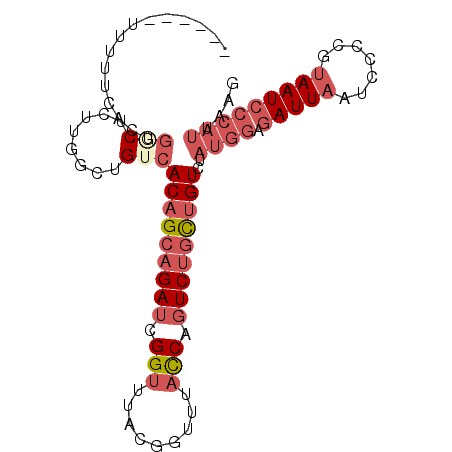
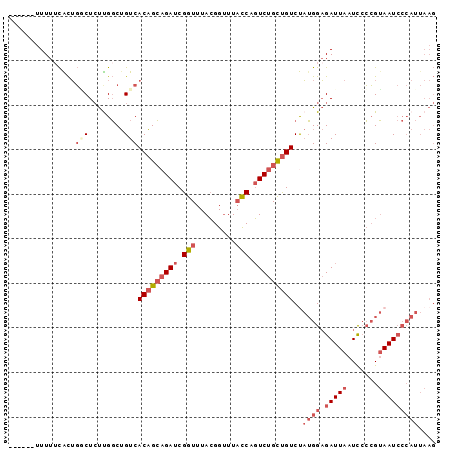
| Location | 12,745,570 – 12,745,660 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12745570 90 - 20766785 CUUAAUGGGAUUACGGGAAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCAAGAGCCAGUGAAAAA------ ....(((((((((.......))))).))))..((((((((.((((.........)))).)))))))).......................------ ( -22.80) >DroSec_CAF1 92027 90 - 1 CUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCAAGAGCCAGUGAAAAA------ .(((.(((......(((((....)))))....((((((((.((((.........)))).))))))))............))).)))....------ ( -25.60) >DroSim_CAF1 99292 90 - 1 CUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCAAGAGCCAGUGAAAAA------ .(((.(((......(((((....)))))....((((((((.((((.........)))).))))))))............))).)))....------ ( -25.60) >DroEre_CAF1 93241 91 - 1 CUUAAUGGGAUUACGGGGAUUAAUCUCCAUGGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCGAGAGUCAGCGAAAAAA----- ........((((.((((((......))).((.((((((((.((((.........)))).))))))))..)).)))..))))..........----- ( -25.40) >DroYak_CAF1 99898 96 - 1 CUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCAAGAGGCAGUGCAAAAAAAGAA ....(((((((((.......))))).))))..((((((((.((((.........)))).))))))))....(((....)))............... ( -27.30) >DroAna_CAF1 86286 87 - 1 ---------AUUUCCUCGAUUAAUCGCCUGGGACGACGGAAGGAAAACCCCUAAACCGAUCUUCAGUGACUGACAGCAGAACACAAAAAAAAAAAA ---------...(((((((....)))...)))).(((((.(((......)))...))).))....(((.(((....)))..)))............ ( -13.60) >consensus CUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGACAGCCAAGAGCCAGUGAAAAA______ .....(((......(((((....)))))....((((((((.((((.........)))).)))))))).....)))..................... (-17.42 = -18.90 + 1.48)

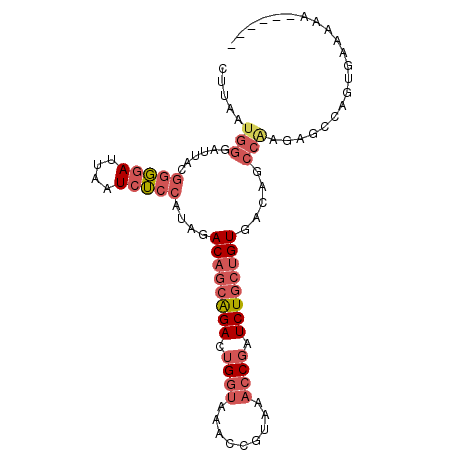
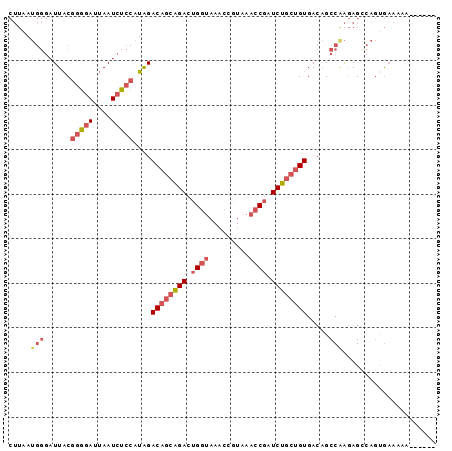
| Location | 12,745,590 – 12,745,700 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -22.72 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12745590 110 + 20766785 GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUUCCCGUAAUCCCAUUAAGAAGCGAAACGAAAAUAUUCCAAACGAAAUGCGAGGACAUU (((.((((((((.(((.........))).))))))))(((((((.(((((.......)))))))))..))).(((...((.............))...)))...)))... ( -26.52) >DroSec_CAF1 92047 110 + 1 GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAGAAGCGAAACGAAAAUAUUCCAAACGAAAUGCGAGGACACU (((.((((((((.(((.........))).))))))))(((((((.(((((.......)))))))))..))).(((...((.............))...)))...)))... ( -26.52) >DroSim_CAF1 99312 110 + 1 GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAGAAGCGAAACGAAAAUAUUCCAAACGAAAUACGAGGACACU (((.((((((((.(((.........))).))))))))(((((((.(((((.......)))))))))..))).................................)))... ( -26.20) >DroEre_CAF1 93262 101 + 1 GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCCAUGGAGAUUAAUCCCCGUAAUCCCAUUAAGAAGGGUAUCGAAAAUAUUCCCCAGGAAAUGC--------- ....((((((((.(((.........))).))))))))(((((((.(((((.......)))))))))......(((....(((....)))))).))).....--------- ( -27.90) >DroYak_CAF1 99924 110 + 1 GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAGAAGGGAGGCGAACAUAUGCCAAAGGAAAUGCGAGAAAAUU ...(((((((((.(((.........))).)))))))))....(((......))).((((.((((........))))((((......))))........))))........ ( -31.90) >consensus GUCACAGCAGAUCGGUUUACGGUUUACCAGUCUGCUGUCUAUGGAGAUUAAUCCCCGUAAUCCCAUUAAGAAGCGAAACGAAAAUAUUCCAAACGAAAUGCGAGGACACU .(((((((((((.(((.........))).)))))))))..((((.(((((.......))))))))).............))............................. (-22.72 = -22.72 + 0.00)

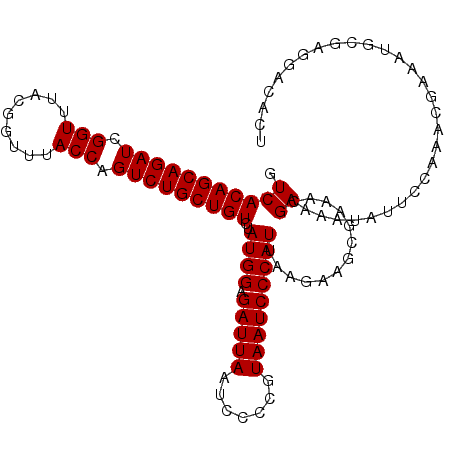
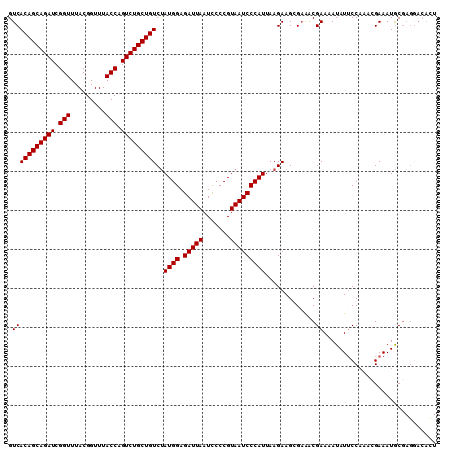
| Location | 12,745,590 – 12,745,700 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12745590 110 - 20766785 AAUGUCCUCGCAUUUCGUUUGGAAUAUUUUCGUUUCGCUUCUUAAUGGGAUUACGGGAAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ...(((.............((((..(((..(((.((.(........).))..)))..))).....))))...((((((((.((((.........)))).))))))))))) ( -28.70) >DroSec_CAF1 92047 110 - 1 AGUGUCCUCGCAUUUCGUUUGGAAUAUUUUCGUUUCGCUUCUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ...(((((((.((..((((.((((..............)))).))))..))..)))))))............((((((((.((((.........)))).))))))))... ( -31.34) >DroSim_CAF1 99312 110 - 1 AGUGUCCUCGUAUUUCGUUUGGAAUAUUUUCGUUUCGCUUCUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ...((((((((((..((((.((((..............)))).))))..).)))))))))............((((((((.((((.........)))).))))))))... ( -34.34) >DroEre_CAF1 93262 101 - 1 ---------GCAUUUCCUGGGGAAUAUUUUCGAUACCCUUCUUAAUGGGAUUACGGGGAUUAAUCUCCAUGGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ---------......((((((((..(((..((...(((........)))....))..)))...)))))).))((((((((.((((.........)))).))))))))... ( -36.10) >DroYak_CAF1 99924 110 - 1 AAUUUUCUCGCAUUUCCUUUGGCAUAUGUUCGCCUCCCUUCUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ....................(((........)))((((........))))....(((((....)))))....((((((((.((((.........)))).))))))))... ( -31.70) >consensus AAUGUCCUCGCAUUUCGUUUGGAAUAUUUUCGUUUCGCUUCUUAAUGGGAUUACGGGGAUUAAUCUCCAUAGACAGCAGACUGGUAAACCGUAAACCGAUCUGCUGUGAC ...........((..((((.((((..............)))).))))..))...(((((....)))))....((((((((.((((.........)))).))))))))... (-23.76 = -24.76 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:46 2006