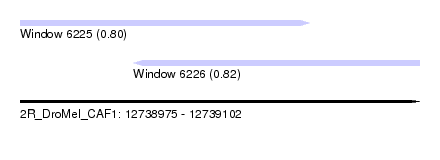
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,738,975 – 12,739,102 |
| Length | 127 |
| Max. P | 0.818702 |

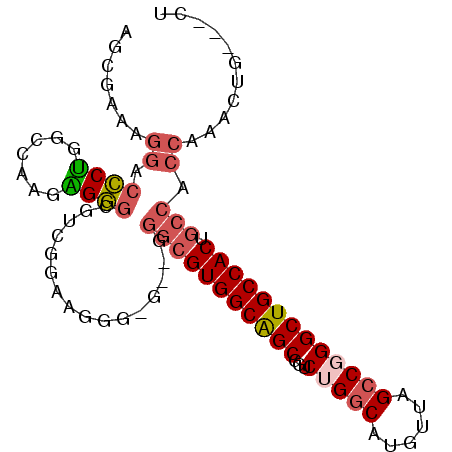
| Location | 12,738,975 – 12,739,067 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -40.64 |
| Consensus MFE | -25.82 |
| Energy contribution | -27.77 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12738975 92 + 20766785 -CGCCACCAGUCGCGUGAGUUCGCGUCAGUUGGCAACAG---CAGUUUGGUGGCAGUGGCAGCCCGGCUAACAUGCCAGACCGCUGCC-ACG-CCC-------C-C -.((((((((.((((......)))).).((((....)))---)....))))))).(((((((((.(((......))).)...))))))-)).-...-------.-. ( -47.80) >DroSec_CAF1 85296 93 + 1 -CGGCACCAGUCGCGUGAGUUCGCGUCAGUUGGCAACAG---CAGUUUGGUGGCAGUGGCAGCCCGGCUAACAUGCCAGACCGCUGCC-ACG-CCC-------CCC -.(.((((((.((((......)))).).((((....)))---)....))))).).(((((((((.(((......))).)...))))))-)).-...-------... ( -43.20) >DroEre_CAF1 86063 92 + 1 -CGCCACCAGUCGCGUGAGUUCGCGUCAGUUGGCAACAG---CAGUUUGGUGGCAGUGGCAGCCGGGCUAACAUGCCAGACCGCUGCC-ACG-CCC-------C-C -.((((((((.((((......)))).).((((....)))---)....))))))).((((((((..(((......))).....))))))-)).-...-------.-. ( -48.80) >DroYak_CAF1 92963 97 + 1 -CGCCACCAGUCGCGUGAGUUCGCGUCAGUUGGCAACAGCAGCAGUUUGGUGGCAGUGGCAGCCCGGCUAACAUGCCAGACCGCUGCC-ACG-CCCCC-----C-C -.((((((((.((((......)))).).((((....)))).......))))))).(((((((((.(((......))).)...))))))-)).-.....-----.-. ( -47.80) >DroAna_CAF1 79395 82 + 1 -UGCCACCAGUCGCAUGAGUUCGCGUCAGUUGGCAACAG---C-----------AGUGGCAGCCUGGCCAACAUGUCUGACCGCCGCC-ACG-CCU-------UCC -((((((..(.(((........))).).((((....)))---)-----------.))))))((.((((.................)))-).)-)..-------... ( -26.43) >DroPer_CAF1 75051 101 + 1 CUGCCACCAGUCACAUCAGUUCGCGUCAGUUGGCAACAG---CAAC--GGCAGCAGCAGCAGCUUGGCCAACAUGCCAGACCGCUAUCCGCAGCCCUCUGCAGCCC (((((....((.((....))..))....((((....)))---)...--)))))..(((((.(..((((......))))..).)))....((((....)))).)).. ( -29.80) >consensus _CGCCACCAGUCGCGUGAGUUCGCGUCAGUUGGCAACAG___CAGUUUGGUGGCAGUGGCAGCCCGGCUAACAUGCCAGACCGCUGCC_ACG_CCC_______C_C ..((((((.(.((((......)))).)...((....))..........)))))).((((((((..(((......))).....)))))).))............... (-25.82 = -27.77 + 1.95)

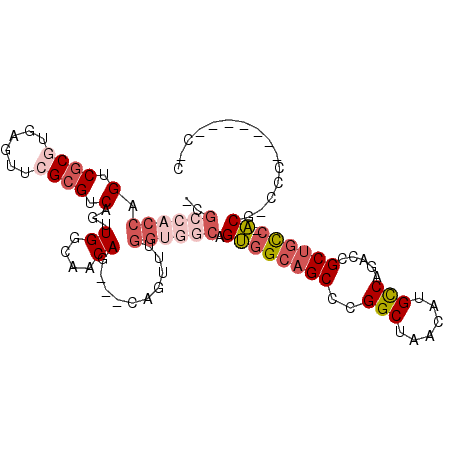
| Location | 12,739,011 – 12,739,102 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -28.17 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12739011 91 - 20766785 AACGAAAGGACCCUGGCCAAGAGGGCGUCGAAAUGG-G--GGGCGUGGCAGCGGUCUGGCAUGUUAGCCGGGCUGCCACUGCCACCAAACUG---CU ..(....)((((((........))).)))....(((-.--.(((((((((((...(((((......))))))))))))).))).))).....---.. ( -40.40) >DroSec_CAF1 85332 92 - 1 AACGAAAGGACCCUGGCCAAGAGGGCGUUGGAAGGGGG--GGGCGUGGCAGCGGUCUGGCAUGUUAGCCGGGCUGCCACUGCCACCAAACUG---CU ..(....)..((((..((((.......))))..))))(--((((((((((((...(((((......))))))))))))).))).))......---.. ( -44.30) >DroSim_CAF1 92586 90 - 1 AGCGAAAGGACCCUGGCCAAGAGGGCGUCGGAAGGG----GGGCGUGGCAGCGGUCUGGCAUGUUAGCCGGGCUGCCACUGCCACCAAACUG---CU ((((...((.(((..(((.....)))..(....)))----)(((((((((((...(((((......))))))))))))).))).))....))---)) ( -42.20) >DroEre_CAF1 86099 90 - 1 -GCGAAAGGACCCUGGCCAAGGGGGCGUCGGAAUGG-G--GGGCGUGGCAGCGGUCUGGCAUGUUAGCCCGGCUGCCACUGCCACCAAACUG---CU -.(((.....((((.....))))....)))...(((-.--.(((((((((((((.((((....)))).)).)))))))).))).))).....---.. ( -39.90) >DroYak_CAF1 92999 96 - 1 AGCGAAAGGACCCUGGCCAAGGGGGCGUCGGAAUGG-GGGGGGCGUGGCAGCGGUCUGGCAUGUUAGCCGGGCUGCCACUGCCACCAAACUGCUGCU ..(....)..((((.....))))((((.(((..(((-....(((((((((((...(((((......))))))))))))).))).)))..))).)))) ( -45.80) >DroAna_CAF1 79431 76 - 1 G--GUCCGGACUCCCGACUCCGGAC---CGGAGGCGGA--AGGCGUGGCGGCGGUCAGACAUGUUGGCCAGGCUGCCACU-----------G---CU (--(((((((........)))))))---)...(.(...--.).)((((((((((((((.....))))))..)))))))).-----------.---.. ( -40.50) >consensus AGCGAAAGGACCCUGGCCAAGAGGGCGUCGGAAGGG_G__GGGCGUGGCAGCGGUCUGGCAUGUUAGCCGGGCUGCCACUGCCACCAAACUG___CU .......((.((((.......))))................(((((((((((...(((((......))))))))))))).))).))........... (-28.17 = -28.87 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:39 2006