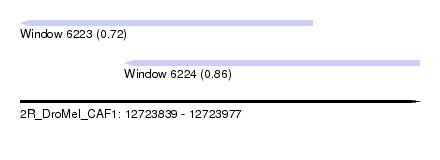
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,723,839 – 12,723,977 |
| Length | 138 |
| Max. P | 0.855750 |

| Location | 12,723,839 – 12,723,940 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -19.35 |
| Energy contribution | -21.60 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
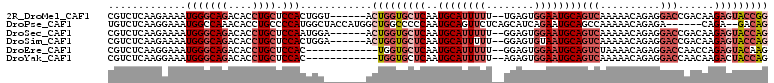
>2R_DroMel_CAF1 12723839 101 - 20766785 ----ACUGGUGCUCAAUGCAUUUUU--UGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCGGUAUCAAGGAGAUGUGAUAAAAAUUAACGACUCCCCG ----((((((((((..((((((((.--.....))))))))(((..............)))..)))))))))).....((((.(((...........))).))))... ( -31.34) >DroPse_CAF1 74104 99 - 1 CAUGGCUGGCCCCCAAUGCAGUUCUCAGCAUCAGAAUGCAGCCAAAAACAGAGA------CAGA--GACAGGAGAGAAGAGAAGUGAUAAAAAUUAACGACUCCCAU ..((((((.......((((........)))).......))))))..........------....--....((.(((..(.....((((....)))).)..))))).. ( -16.64) >DroSec_CAF1 70156 101 - 1 ----ACUGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAGUAUCAGGGAGAUGUGAUAAAAAUUAACGACUCCCCG ----((((((((((..((((((((.--.....))))))))(((..............)))..))))))))))....(((((.(((...........))).))))).. ( -35.34) >DroSim_CAF1 76018 101 - 1 ----ACUGGUGCUCAAUGCAUUUUU--GGAGUGUAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAGUAUCAAGGAGAUGUGAUAAAAAUUAACGACUCCCCG ----((((((((((..((((((...--.......))))))(((..............)))..)))))))))).....((((.(((...........))).))))... ( -29.14) >DroEre_CAF1 71004 99 - 1 ------UGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCUAAAACAGAGGACCAACCAGAGUACAAGUAUCAAGGAGAUGUGAUAAAAAUUAACGACUCCCCA ------..((((((..((((((((.--.....))))))))((((........))))......)))))).........((((.(((...........))).))))... ( -25.30) >DroYak_CAF1 77910 99 - 1 ------UGGUGCUCAAUGCAUUUUU--AGAGUGGAAUGCAGUCAAAAACAGAGGACCAACAAGACUACCAGUAUCAAGGAGAUGUGAUAAAAAUUAACGACUCCCCA ------((((.(((..((((((((.--.....))))))))((.....)).))).))))...................((((.(((...........))).))))... ( -22.90) >consensus ____ACUGGUGCUCAAUGCAUUUUU__GGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAGUAUCAAGGAGAUGUGAUAAAAAUUAACGACUCCCCG .....(((((((((..((((((((........))))))))(((..........)))......)))))))))......((((.(((...........))).))))... (-19.35 = -21.60 + 2.25)

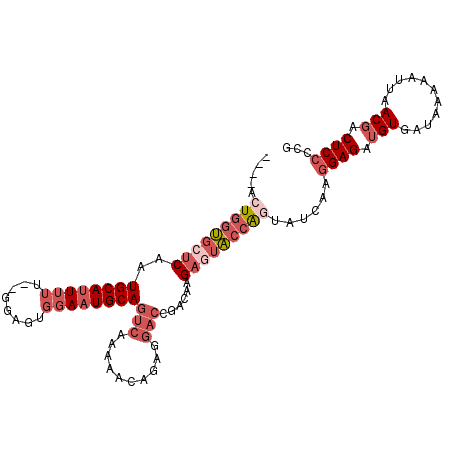
| Location | 12,723,875 – 12,723,977 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -20.85 |
| Energy contribution | -23.10 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12723875 102 - 20766785 CGUCUCAAGAAAAUGGGCAGACACCUGCUCCACUGGU------ACUGGUGCUCAAUGCAUUUUU--UGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCGG .............(((((((....)))).)))(((((------((((((.(((..((((((((.--.....))))))))((.....)).))).)))......)))))))) ( -33.30) >DroPse_CAF1 74140 102 - 1 UGUCUCAAGGAAAUGGCCAAACACCUGCCCCAUGGCUACCAUGGCUGGCCCCCAAUGCAGUUCUCAGCAUCAGAAUGCAGCCAAAAACAGAGA------CAGA--GACAG ((((((.......((((.........(((((((((...))))))..)))......((((.((((.......))))))))))))......))))------))..--..... ( -28.72) >DroSec_CAF1 70192 102 - 1 CGUCUCAAGAAAAUGGGCAGACACCUGCUCCAAUGGA------ACUGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAG .............(((((((....)))).))).....------.(((((((((..((((((((.--.....))))))))(((..............)))..))))))))) ( -33.04) >DroSim_CAF1 76054 102 - 1 CGUCUCAAGAAAAUGGGCAGACACCUGCUCCACUGGA------ACUGGUGCUCAAUGCAUUUUU--GGAGUGUAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAG .............(((((((....)))).))).....------.(((((((((..((((((...--.......))))))(((..............)))..))))))))) ( -29.84) >DroEre_CAF1 71040 96 - 1 CGUCUCAAGGAAAUGGGCAGACACCUGCUCCAC------------UGGUGCUCAAUGCAUUUUU--GGAGUGGAAUGCAGUCUAAAACAGAGGACCAACCAGAGUACAAG .(((((..((...(((((((....)))).))).------------((((.(((..((((((((.--.....))))))))((.....)).))).)))).)).))).))... ( -30.80) >DroYak_CAF1 77946 96 - 1 CGUCUCAAGGAAAUGGGCAGACACCUGCUCCAC------------UGGUGCUCAAUGCAUUUUU--AGAGUGGAAUGCAGUCAAAAACAGAGGACCAACAAGACUACCAG .((((........(((((((....)))).))).------------((((.(((..((((((((.--.....))))))))((.....)).))).))))...))))...... ( -29.10) >consensus CGUCUCAAGAAAAUGGGCAGACACCUGCUCCACUGG_______ACUGGUGCUCAAUGCAUUUUU__GGAGUGGAAUGCAGUCAAAAACAGAGGACCGACAAGAGUACCAG .............(((((((....)))).)))............(((((((((..((((((((........))))))))(((..........)))......))))))))) (-20.85 = -23.10 + 2.25)
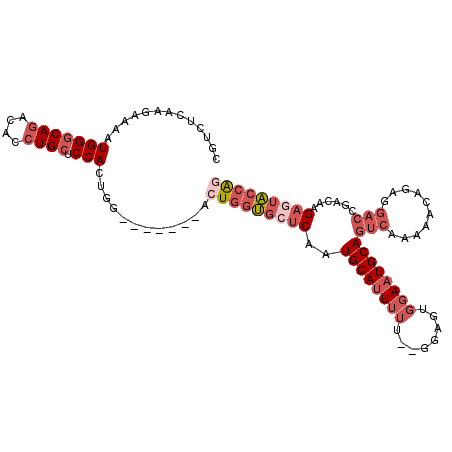
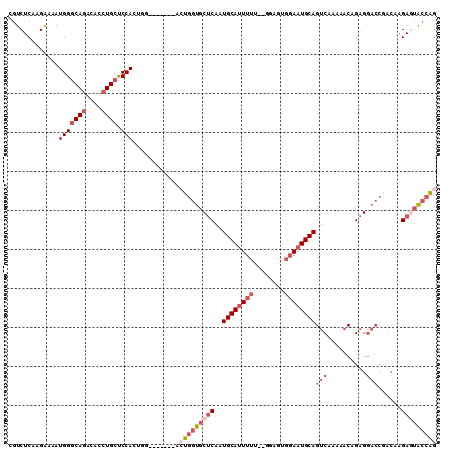
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:37 2006