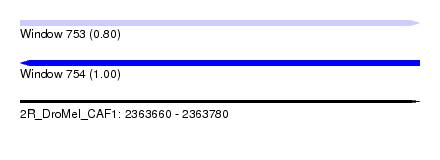
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,363,660 – 2,363,780 |
| Length | 120 |
| Max. P | 0.995818 |

| Location | 2,363,660 – 2,363,780 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.92 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2363660 120 + 20766785 UAACGGGGCAUUAGAGCUGCUUAGCAGCCACUGUGUGACCUCGGAGUCUGGCAUGUCCCCUCCAUGGCUGCUAAGUCCGCUUGAGCAUUCGACUAAGCCCAUUUAAUUGUCUUCAUUAGC .....((((.((((....((((((((((((......(....)((((...((.....)).)))).))))))))))))..((....))......)))))))).................... ( -37.10) >DroSec_CAF1 13181 120 + 1 UAACGGGGCAUUAGAGCUGCUUAGCAGCCACUGUGUGGCCUCGGAGUCUGGCAUGUCCCCUCCAUGGCUGCUAAGUCCGCUUGAGCAUUCGCCUAAGCCCAUUUAAUUGUCUUCAUUAGC .....((((.((((.((((((((((.(((((...)))))...(((.(.(((((.(((........))))))))).))))).))))))...)))))))))).................... ( -40.10) >DroSim_CAF1 13285 120 + 1 UAACGGGGCAUUAGAGCUGCUUAGCAGCCACUGUGUGGCCUCGGAGUCUGGCAUGUCCCCUCCAUGGCUGCUAAGUCCGCUUGAGCAUUCGCCUAAGCCCAUUUAAUUGUCUUCAUUAGC .....((((.((((.((((((((((.(((((...)))))...(((.(.(((((.(((........))))))))).))))).))))))...)))))))))).................... ( -40.10) >DroEre_CAF1 12943 109 + 1 UAAAGGGGCUUUAGAGCUGCUUAACAGCCACUGUGUGGCCUCGGAGUC----------UCUCCAUGGUAGCUAAGUCCGCUUGAGCAUUC-CCUAAGCCCAUUUAAUUGUCUUCAUUAGC ((((.((((((.(((((((((.....(((((...)))))...((((..----------.))))..)))))))......((....))....-.)))))))).))))............... ( -32.60) >DroYak_CAF1 12668 120 + 1 UAAUGGGGCAUUAGAGCUGCUUAACAGCCACUGUGUGGCCUCGGAGUCUGGGAAGUCCUCGCCAUGGCUGCUAAGUCAGCUAGAGCAUUCGCCUAAGCCCAUUUAAUUGUCUUCAUUAAC .....((((.(((((((((((((.((((((..(((.((((((((...)))))..).)).)))..)))))).)))).)))))...((....)))))))))).................... ( -41.40) >consensus UAACGGGGCAUUAGAGCUGCUUAGCAGCCACUGUGUGGCCUCGGAGUCUGGCAUGUCCCCUCCAUGGCUGCUAAGUCCGCUUGAGCAUUCGCCUAAGCCCAUUUAAUUGUCUUCAUUAGC .....((((.((((.((.(((((((((((((.....).....((((...((.....)).)))).))))))))))))..((....))....)))))))))).................... (-31.78 = -32.92 + 1.14)

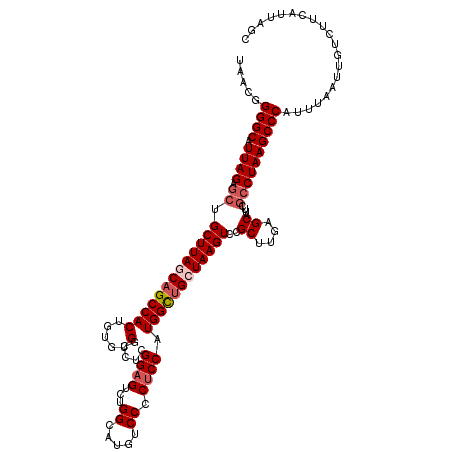
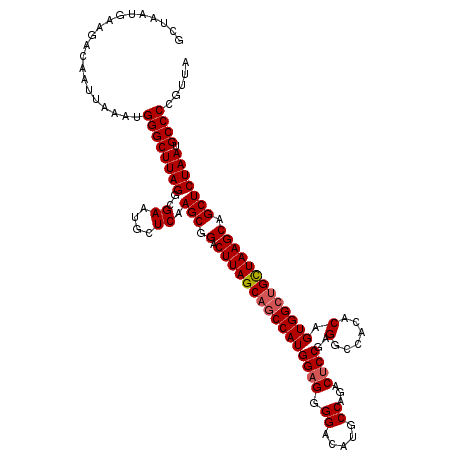
| Location | 2,363,660 – 2,363,780 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -35.00 |
| Energy contribution | -35.46 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2363660 120 - 20766785 GCUAAUGAAGACAAUUAAAUGGGCUUAGUCGAAUGCUCAAGCGGACUUAGCAGCCAUGGAGGGGACAUGCCAGACUCCGAGGUCACACAGUGGCUGCUAAGCAGCUCUAAUGCCCCGUUA ....................((((((((..((....)).(((.(.((((((((((((((((.((.....))...))))..(......).))))))))))))).))))))).))))..... ( -40.40) >DroSec_CAF1 13181 120 - 1 GCUAAUGAAGACAAUUAAAUGGGCUUAGGCGAAUGCUCAAGCGGACUUAGCAGCCAUGGAGGGGACAUGCCAGACUCCGAGGCCACACAGUGGCUGCUAAGCAGCUCUAAUGCCCCGUUA ....................((((((((((...(((....)))(.((((((((((((((((.((.....))...))))..(......).))))))))))))).)).)))).))))..... ( -43.40) >DroSim_CAF1 13285 120 - 1 GCUAAUGAAGACAAUUAAAUGGGCUUAGGCGAAUGCUCAAGCGGACUUAGCAGCCAUGGAGGGGACAUGCCAGACUCCGAGGCCACACAGUGGCUGCUAAGCAGCUCUAAUGCCCCGUUA ....................((((((((((...(((....)))(.((((((((((((((((.((.....))...))))..(......).))))))))))))).)).)))).))))..... ( -43.40) >DroEre_CAF1 12943 109 - 1 GCUAAUGAAGACAAUUAAAUGGGCUUAGG-GAAUGCUCAAGCGGACUUAGCUACCAUGGAGA----------GACUCCGAGGCCACACAGUGGCUGUUAAGCAGCUCUAAAGCCCCUUUA ...............((((.(((((((((-(..(((....)))(.((((((.....(((((.----------..))))).((((((...)))))))))))))..)))).)))))).)))) ( -33.40) >DroYak_CAF1 12668 120 - 1 GUUAAUGAAGACAAUUAAAUGGGCUUAGGCGAAUGCUCUAGCUGACUUAGCAGCCAUGGCGAGGACUUCCCAGACUCCGAGGCCACACAGUGGCUGUUAAGCAGCUCUAAUGCCCCAUUA (((......)))........((((((((((....))...(((((.((((((((((((.(.(.((.((((.........)))))).).).))))))))))))))))))))).))))..... ( -42.90) >consensus GCUAAUGAAGACAAUUAAAUGGGCUUAGGCGAAUGCUCAAGCGGACUUAGCAGCCAUGGAGGGGACAUGCCAGACUCCGAGGCCACACAGUGGCUGCUAAGCAGCUCUAAUGCCCCGUUA ....................((((((((..((....)).(((.(.((((((((((((((((.((.....))...))))..(......).))))))))))))).))))))).))))..... (-35.00 = -35.46 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:06 2006