| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,714,763 – 12,714,853 |
| Length | 90 |
| Max. P | 0.564798 |

| Location | 12,714,763 – 12,714,853 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.70 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
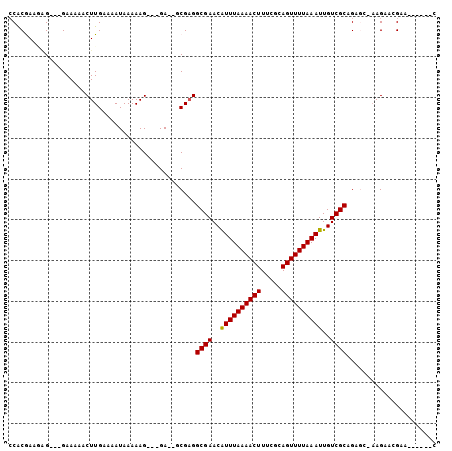
>2R_DroMel_CAF1 12714763 90 + 20766785 CCACGAAGUG---GAAAGACUUGAAAAUAAAAAG---GA--GCGAGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCAGAGC-AAGAAGGAA------C ((((...)))---)....................---..--((...((((..((((((((((.....))))))))))..))))...))-.........------. ( -19.90) >DroPse_CAF1 66227 98 + 1 CCACAAA-AG---GAAAAACUUGAAAAUAAAAAGAACGA--ACGAGGCGAACGUUUAAAACUUUCGCAGUUUUAAAUUGUCGCCGCAA-AAGAACGAAACGAAAC ((.....-.)---).........................--..(.(((((..((((((((((.....))))))))))..))))).)..-................ ( -17.60) >DroEre_CAF1 61960 90 + 1 CCGCGAAGAG---GAAAGACUUGAAAAUAAAAAG---GA--GCGAGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCAGAGG-AAGAAGGAA------U .(((...(((---......)))............---..--)))..((((..((((((((((.....))))))))))..)))).....-.........------. ( -16.13) >DroYak_CAF1 64527 92 + 1 CCACGAAGAG---GAAAGACUUGAAAAUAAAAAG---GAACGCGAGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCAGCGC-AAGUAGGAA------U ((((...(((---......)))............---....(((..((((..((((((((((.....))))))))))..))))..)))-..)).))..------. ( -23.00) >DroAna_CAF1 57406 94 + 1 ACACGAAAGGAAAGAAAAACUUGAAAAUAAAAAG---GA--GCGUGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCCGAGCCCAGGACGAA------U ...(....)........................(---(.--((.((((((..((((((((((.....))))))))))..)))))).))))........------. ( -24.50) >DroPer_CAF1 56441 98 + 1 CCACAAA-AG---GAAAAACUUGAAAAUAAAAAGAACGA--ACGAGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCCGCAA-AAGAACGAAAGGAAAC ((....(-((---......))).................--..(.(((((..((((((((((.....))))))))))..))))).)..-..........)).... ( -19.00) >consensus CCACGAAGAG___GAAAAACUUGAAAAUAAAAAG___GA__GCGAGGCGAACAUUUAAAACUUUCGCAGUUUUAAAUUGUCGCAGAGC_AAGAACGAA______C ..............................................((((..((((((((((.....))))))))))..))))...................... (-12.84 = -12.70 + -0.14)


| Location | 12,714,763 – 12,714,853 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -13.08 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
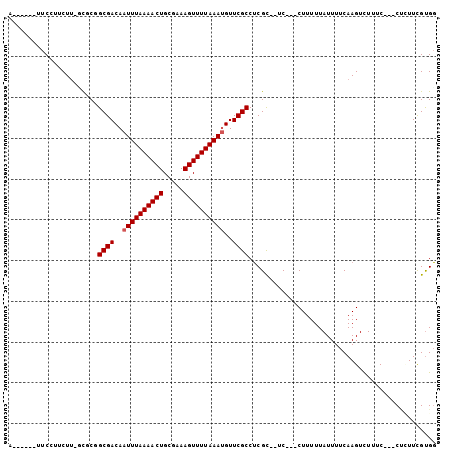
>2R_DroMel_CAF1 12714763 90 - 20766785 G------UUCCUUCUU-GCUCUGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCUCGC--UC---CUUUUUAUUUUCAAGUCUUUC---CACUUCGUGG .------.........-((...((((..((((((((((.....))))))))))..))))...))--..---....................(---(((...)))) ( -17.40) >DroPse_CAF1 66227 98 - 1 GUUUCGUUUCGUUCUU-UUGCGGCGACAAUUUAAAACUGCGAAAGUUUUAAACGUUCGCCUCGU--UCGUUCUUUUUAUUUUCAAGUUUUUC---CU-UUUGUGG ....(((..(((....-..))))))((((...((((((..((((((....((((..((...)).--.))))......)))))).))))))..---..-.)))).. ( -15.90) >DroEre_CAF1 61960 90 - 1 A------UUCCUUCUU-CCUCUGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCUCGC--UC---CUUUUUAUUUUCAAGUCUUUC---CUCUUCGCGG .------.........-.....((((..((((((((((.....))))))))))..))))..(((--..---.....................---......))). ( -14.55) >DroYak_CAF1 64527 92 - 1 A------UUCCUACUU-GCGCUGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCUCGCGUUC---CUUUUUAUUUUCAAGUCUUUC---CUCUUCGUGG .------...((((..-(((..((((..((((((((((.....))))))))))..))))..)))....---............(((......---..))).)))) ( -20.40) >DroAna_CAF1 57406 94 - 1 A------UUCGUCCUGGGCUCGGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCACGC--UC---CUUUUUAUUUUCAAGUUUUUCUUUCCUUUCGUGU .------........((((..(((((..((((((((((.....))))))))))..)))))..))--))---.................................. ( -22.20) >DroPer_CAF1 56441 98 - 1 GUUUCCUUUCGUUCUU-UUGCGGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCUCGU--UCGUUCUUUUUAUUUUCAAGUUUUUC---CU-UUUGUGG ................-..(.(((((..((((((((((.....))))))))))..))))).)..--..........................---..-....... ( -17.80) >consensus A______UUCCUUCUU_GCGCGGCGACAAUUUAAAACUGCGAAAGUUUUAAAUGUUCGCCUCGC__UC___CUUUUUAUUUUCAAGUCUUUC___CUCUUCGUGG ......................((((..((((((((((.....))))))))))..)))).............................................. (-13.08 = -13.25 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:30 2006