
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,710,992 – 12,711,112 |
| Length | 120 |
| Max. P | 0.926639 |

| Location | 12,710,992 – 12,711,112 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -28.76 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12710992 120 + 20766785 CCCUGAUCAGAAAAGAACUGAAUAGCUGUUUGGCCAAUUCGGUCAUCUGUUUGAAACCGGCAUUCAAACGAUCUAUGGGCGAACGCUCACUGCUGCAAAACUACUGCAACAUAUUUGUAA ....((((.....((((((((((.(((....)))..)))))))..)))(((((((.......)))))))))))..(((((....))))).((.((((.......)))).))......... ( -28.20) >DroSec_CAF1 57295 120 + 1 CACUGAUCAGAAAUGAACUGAAUAGCUGUUUGGCCGAUUCGGUCAUCUCUUUGAAACCGGCAUUCAAACGAGCUAUGGGCGAACGCUCACUUUUGCAAAACUACUGCAACAUAUUUGUAA .......((((.(((......(((((((((((((((.(((((........)))))..))))...))))).))))))((((....))))....(((((.......)))))))).))))... ( -29.70) >DroSim_CAF1 63295 120 + 1 CACUGAUCAGAAAUGAACUGAAUAGCUGUUUGGCCGAUUCGGUCAUCUCUUUGAAACCGGCAUUCAAACGAGCUAUGGGCGAUCGCUCACUGUUGCAAAACUACUGCAACAUAUUUGUAA ......(((....))).....(((((((((((((((.(((((........)))))..))))...))))).))))))((((....))))..(((((((.......)))))))......... ( -33.80) >DroEre_CAF1 58130 120 + 1 CGCUGGUCAGAGUUGAUCUGAAUGGUCGUUUGGCUGACUUGGUCAUCUCCCUAAAGCCGGCAUUCAAACGCUCCAUGGGCGAAAGCCCAUUGUUGCAAAACUAUUGCAACAUAUUUGUGA .((((((..(((.(((((.....(((((......))))).))))).)))......)))))).......(((...((((((....))))))((((((((.....)))))))).....))). ( -43.80) >DroYak_CAF1 60613 120 + 1 CACUGGUUAGAACUAAUCUGGAUGGGCGUUUGGCCGAUUUGGUCAUCUCCUUGAAACCGGCAUUCAAACGCUCCAUGGGCGACAACCCACUGUUGCAAAACUAUUGCAACAUAUUUGUAA ..(.(((((....))))).).(((((((((((((((......(((......)))...))))...))))))).))))(((......)))..((((((((.....))))))))......... ( -35.80) >consensus CACUGAUCAGAAAUGAACUGAAUAGCUGUUUGGCCGAUUCGGUCAUCUCUUUGAAACCGGCAUUCAAACGAUCUAUGGGCGAACGCUCACUGUUGCAAAACUACUGCAACAUAUUUGUAA ......((((.......))))(((((((((((((((..((((........))))...))))...)))))).)))))((((....))))..(((((((.......)))))))......... (-28.76 = -27.80 + -0.96)



| Location | 12,710,992 – 12,711,112 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -26.24 |
| Energy contribution | -25.04 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12710992 120 - 20766785 UUACAAAUAUGUUGCAGUAGUUUUGCAGCAGUGAGCGUUCGCCCAUAGAUCGUUUGAAUGCCGGUUUCAAACAGAUGACCGAAUUGGCCAAACAGCUAUUCAGUUCUUUUCUGAUCAGGG .........((((((((.....))))))))(((.((....)).))).(((((((((((.......)))))))(((.(((.((((.(((......))))))).)))....))))))).... ( -31.30) >DroSec_CAF1 57295 120 - 1 UUACAAAUAUGUUGCAGUAGUUUUGCAAAAGUGAGCGUUCGCCCAUAGCUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCGAAUCGGCCAAACAGCUAUUCAGUUCAUUUCUGAUCAGUG .(((.......((((((.....))))))((((((((........((((((.(((((...((((((((((......)))...))))))))))))))))))...)))))))).......))) ( -30.30) >DroSim_CAF1 63295 120 - 1 UUACAAAUAUGUUGCAGUAGUUUUGCAACAGUGAGCGAUCGCCCAUAGCUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCGAAUCGGCCAAACAGCUAUUCAGUUCAUUUCUGAUCAGUG .........((((((((.....))))))))((((....))))..((((((.(((((...((((((((((......)))...))))))))))))))))))((((.......))))...... ( -34.00) >DroEre_CAF1 58130 120 - 1 UCACAAAUAUGUUGCAAUAGUUUUGCAACAAUGGGCUUUCGCCCAUGGAGCGUUUGAAUGCCGGCUUUAGGGAGAUGACCAAGUCAGCCAAACGACCAUUCAGAUCAACUCUGACCAGCG .........((((((((.....))))))))((((((....))))))((..((((((...((.(((((..((.......))))))).)))))))).))..(((((.....)))))...... ( -38.40) >DroYak_CAF1 60613 120 - 1 UUACAAAUAUGUUGCAAUAGUUUUGCAACAGUGGGUUGUCGCCCAUGGAGCGUUUGAAUGCCGGUUUCAAGGAGAUGACCAAAUCGGCCAAACGCCCAUCCAGAUUAGUUCUAACCAGUG .........((((((((.....))))))))((((((....))))))((.(((((((...((((((((...((......)))))))))))))))))))....(((.....)))........ ( -40.20) >consensus UUACAAAUAUGUUGCAGUAGUUUUGCAACAGUGAGCGUUCGCCCAUAGAUCGUUUGAAUGCCGGUUUCAAAGAGAUGACCGAAUCGGCCAAACAGCUAUUCAGUUCAUUUCUGAUCAGUG .........((((((((.....)))))))).(((..........((((...(((((...((((((((((......)))...))))))))))))..))))((((.......)))))))... (-26.24 = -25.04 + -1.20)

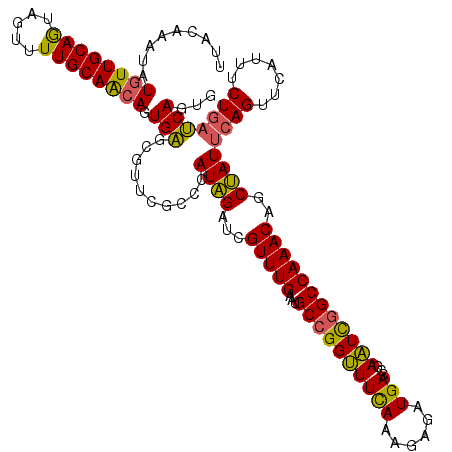
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:29 2006