
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
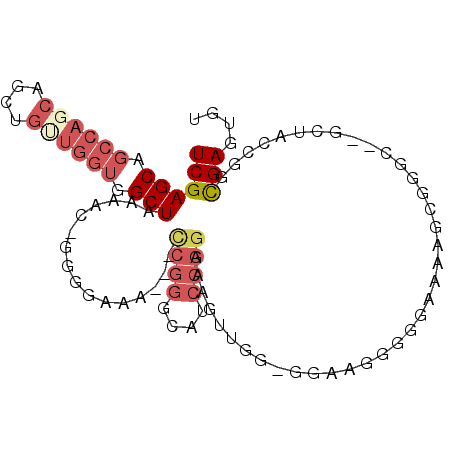
| Location | 12,706,889 – 12,706,983 |
| Length | 94 |
| Max. P | 0.655977 |

| Location | 12,706,889 – 12,706,983 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -13.25 |
| Energy contribution | -14.75 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
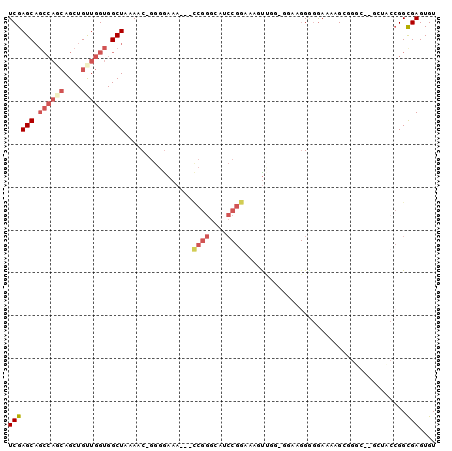
>2R_DroMel_CAF1 12706889 94 + 20766785 UCGAGCAGCCAGCAGCUGUUGGUGGCUAAAAC-GGGGAAA---CCGGGCUUCCGGAAAGUUGACGGAAGGGGAAAAAGCCGUC--GCUACCGGCGAGUGU ...(((.((((((....)))))).)))....(-.((....---)).)((((((((..(((.(((((............)))))--))).)))).)))).. ( -35.60) >DroSec_CAF1 53186 84 + 1 UCGAGCAGCCAGCAGCUGUUGGUGGCUAAAAC-GGGGAAA---CCGGGCAUCCGGCACGUUGGUGGAGGGG-GAAAAGC-----------CGGCGAGUGU ....((.((((((....))))))((((....(-.((....---)).)...(((..(((....)))..))).-....)))-----------).))...... ( -28.50) >DroSim_CAF1 59164 94 + 1 UCGAGCAGCCACCAGCUGUUGGUGGCUAAAAC-GGGGAAA---CCGGGCAUCCGGCACGUUGGUGGAGGGGGGAAAAGCCGUC--GCUACCGGCGAGUGU ....(((((((((((...)))))))))....(-.((....---)).))).....((.(((((((((.((.((......)).))--.))))))))).)).. ( -42.10) >DroEre_CAF1 53881 82 + 1 UCGAGCAGCCAGCAGCUGCUGGUGGCUAAAACCGGGGAAA---CCGGGUAGCCGGAAAAU-------AGGUGGAGGGGAGGGG--------GGUGAGUGU ((.(.(.((((((....))))))(((((...((((.....---)))).))))).......-------.).).)).........--------......... ( -28.50) >DroYak_CAF1 56273 95 + 1 UCGAGCAGCCAGCAGCUGUUGGUGGCUAAAACGGGGGAAA---UCGGGUAACCGGAAAACCCGGAAAAGGGGGAAAAGGGGGA--GCUACCGGCGAGUGU (((((((((.....))))))(((((((....(........---((((....))))....(((......)))......)....)--))))))..))).... ( -30.60) >DroAna_CAF1 52032 81 + 1 UCGAGC-------AGCUGUUUGUGGCUAAAAC-CACAAAAAAAAAGGCCAG--GGCGAGU---------GAGGAAGAGAGGGCAGGAGUCCGGCGAGUGU (((.((-------.(((.(((((((......)-))))))......)))...--.))....---------..........((((....))))..))).... ( -18.80) >consensus UCGAGCAGCCAGCAGCUGUUGGUGGCUAAAAC_GGGGAAA___CCGGGCAUCCGGAAAGUUGG_GGAAGGGGGAAAAGCGGGC__GCUACCGGCGAGUGU ((((((.((((((....)))))).)))................((((....))))......................................))).... (-13.25 = -14.75 + 1.50)

| Location | 12,706,889 – 12,706,983 |
|---|---|
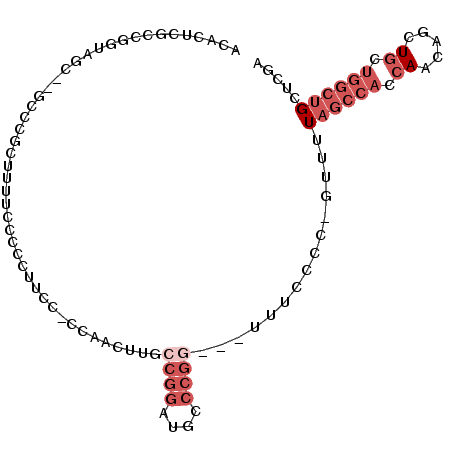
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 70.04 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -6.17 |
| Energy contribution | -8.00 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
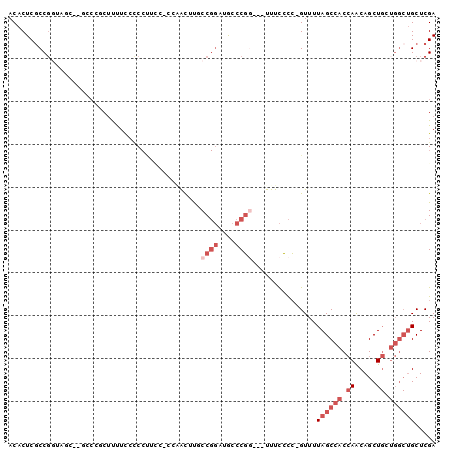
>2R_DroMel_CAF1 12706889 94 - 20766785 ACACUCGCCGGUAGC--GACGGCUUUUUCCCCUUCCGUCAACUUUCCGGAAGCCCGG---UUUCCCC-GUUUUAGCCACCAACAGCUGCUGGCUGCUCGA ......(((((((((--...((((.......((((((.........))))))..(((---.....))-)....)))).......)))))))))....... ( -29.70) >DroSec_CAF1 53186 84 - 1 ACACUCGCCG-----------GCUUUUC-CCCCUCCACCAACGUGCCGGAUGCCCGG---UUUCCCC-GUUUUAGCCACCAACAGCUGCUGGCUGCUCGA ......((((-----------((.....-..........((((.(((((....))))---).....)-)))..(((........)))))))))....... ( -21.80) >DroSim_CAF1 59164 94 - 1 ACACUCGCCGGUAGC--GACGGCUUUUCCCCCCUCCACCAACGUGCCGGAUGCCCGG---UUUCCCC-GUUUUAGCCACCAACAGCUGGUGGCUGCUCGA ......(((((..((--(.((((.....................))))..)))))))---).....(-(...(((((((((.....)))))))))..)). ( -31.30) >DroEre_CAF1 53881 82 - 1 ACACUCACC--------CCCCUCCCCUCCACCU-------AUUUUCCGGCUACCCGG---UUUCCCCGGUUUUAGCCACCAGCAGCUGCUGGCUGCUCGA .........--------................-------.......(((((.((((---.....))))...))))).(.((((((.....)))))).). ( -22.90) >DroYak_CAF1 56273 95 - 1 ACACUCGCCGGUAGC--UCCCCCUUUUCCCCCUUUUCCGGGUUUUCCGGUUACCCGA---UUUCCCCCGUUUUAGCCACCAACAGCUGCUGGCUGCUCGA ......(((((((((--(...........(((......)))......(((((..((.---.......))...)))))......))))))))))....... ( -26.60) >DroAna_CAF1 52032 81 - 1 ACACUCGCCGGACUCCUGCCCUCUCUUCCUC---------ACUCGCC--CUGGCCUUUUUUUUUGUG-GUUUUAGCCACAAACAGCU-------GCUCGA ........((..(..(((.............---------....((.--...)).......((((((-((....)))))))))))..-------)..)). ( -16.50) >consensus ACACUCGCCGGUAGC__GCCCGCUUUUCCCCCUUCC_CCAACUUGCCGGAUGCCCGG___UUUCCCC_GUUUUAGCCACCAACAGCUGCUGGCUGCUCGA .............................................((((....))))...............((((((.((.....)).))))))..... ( -6.17 = -8.00 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:27 2006