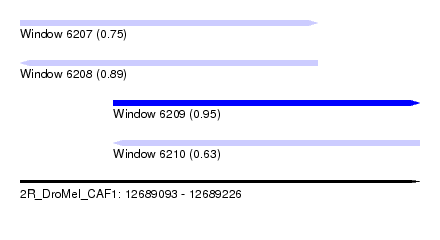
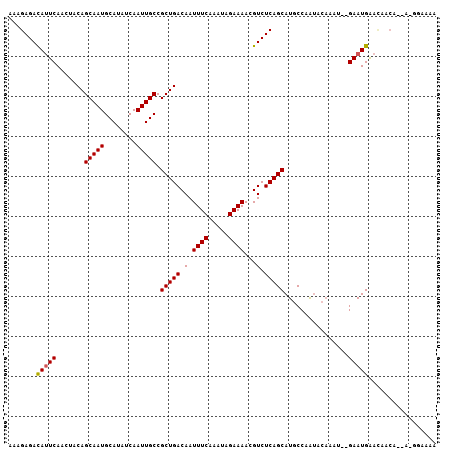
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,689,093 – 12,689,226 |
| Length | 133 |
| Max. P | 0.951031 |

| Location | 12,689,093 – 12,689,192 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -10.16 |
| Energy contribution | -10.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12689093 99 + 20766785 -AGCAG-AG------CCAACCGCAAUCGGUAGACAAAACAAAGAGACAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAG -.....-..------...((((....))))............(((((.........((((...(((.......))).))))....((((.....))))..))))).. ( -19.60) >DroVir_CAF1 62177 97 + 1 GAGAAA-AAAGCAAACCGAACCCAAUCAGUGGAUAAAAAAAA---------AACCACAGCAAUGCAUAUCAAUUGCUGCUGACAAUUUCAAAUAGAAAAUGUCUCAG ..((..-..(((................((((..........---------..))))((((((........)))))))))((((.((((.....)))).)))))).. ( -19.30) >DroPse_CAF1 52510 100 + 1 -AGCAGCAA------CCAACCGCAAUCGGCAGACAAAACAAAGAGACAUUCAGCUGCAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAG -.((.((..------......)).....))............(((((..(((((....(((((........))))).)))))...((((.....))))..))))).. ( -21.00) >DroGri_CAF1 47733 92 + 1 AAGAGA-AG-ACAACUCGAACGCAAUCAGUAGACCAAAAAAA---------AA----AGAAAUGCAUAUCAAUUGCAGCUGACAAUUUCAAAUAGAAAAUGUCUCAG ..(((.-..-....)))((..((..((....)).........---------..----.....((((.......)))))).((((.((((.....)))).)))))).. ( -12.30) >DroAna_CAF1 40993 99 + 1 -AUCAA-AG------CCAACCGCAAUCGGUAGACAAAACAAAGAGACACUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAG -.....-..------...((((....))))............(((((.........((((...(((.......))).))))....((((.....))))..))))).. ( -19.60) >DroPer_CAF1 43117 100 + 1 -AGCAGCAA------CCAACCGCAAUCGGCAGACAAAACAAAGAGACAUUCAGCUGCAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAG -.((.((..------......)).....))............(((((..(((((....(((((........))))).)))))...((((.....))))..))))).. ( -21.00) >consensus _AGCAA_AA______CCAACCGCAAUCGGUAGACAAAACAAAGAGACAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAG ............................(.((((......................((((...(((.......))).))))....((((.....))))..))))).. (-10.16 = -10.22 + 0.06)


| Location | 12,689,093 – 12,689,192 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12689093 99 - 20766785 CUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUGUCUCUUUGUUUUGUCUACCGAUUGCGGUUGG------CU-CUGCU- ..(((((((((.((...(((.....)))((((((((........))))))))))..)))))))))...((...(((.((((....)))).))------).-..)).- ( -28.30) >DroVir_CAF1 62177 97 - 1 CUGAGACAUUUUCUAUUUGAAAUUGUCAGCAGCAAUUGAUAUGCAUUGCUGUGGUU---------UUUUUUUUAUCCACUGAUUGGGUUCGGUUUGCUUU-UUUCUC ((((((((.((((.....)))).)))).((((((((........))))))))....---------........(((((.....)))))))))........-...... ( -21.90) >DroPse_CAF1 52510 100 - 1 CUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGCAGCUGAAUGUCUCUUUGUUUUGUCUGCCGAUUGCGGUUGG------UUGCUGCU- ....((((.((((.....)))).))))(((((((((..((.((((...(.(((((.(((((......))))).).)))).)..))))))..)------))))))))- ( -34.80) >DroGri_CAF1 47733 92 - 1 CUGAGACAUUUUCUAUUUGAAAUUGUCAGCUGCAAUUGAUAUGCAUUUCU----UU---------UUUUUUUGGUCUACUGAUUGCGUUCGAGUUGU-CU-UCUCUU ....((((.((((.....)))).))))....((((((((.(((((..((.----..---------...............)).))))))).))))))-..-...... ( -15.97) >DroAna_CAF1 40993 99 - 1 CUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAGUGUCUCUUUGUUUUGUCUACCGAUUGCGGUUGG------CU-UUGAU- ..((((((.((((.....))))...(((((((((((........))))))))...))).))))))........(((.((((....)))).))------).-.....- ( -24.80) >DroPer_CAF1 43117 100 - 1 CUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGCAGCUGAAUGUCUCUUUGUUUUGUCUGCCGAUUGCGGUUGG------UUGCUGCU- ....((((.((((.....)))).))))(((((((((..((.((((...(.(((((.(((((......))))).).)))).)..))))))..)------))))))))- ( -34.80) >consensus CUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUGUCUCUUUGUUUUGUCUACCGAUUGCGGUUGG______CU_CUGCU_ ....((((.((((.....)))).)))).((((((((........))))))))......................((.((((....)))).))............... (-16.00 = -16.08 + 0.09)

| Location | 12,689,124 – 12,689,226 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
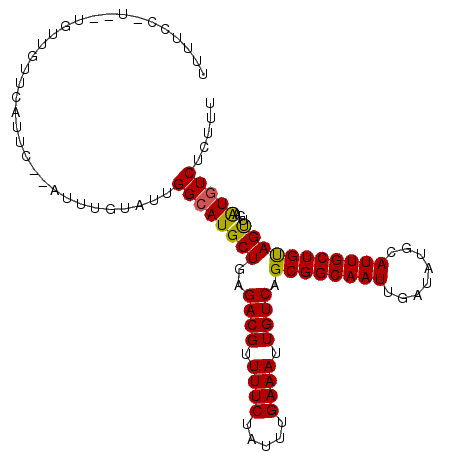
>2R_DroMel_CAF1 12689124 102 + 20766785 AAAGAGACAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAAUACAAAU--GAAUGAACAGCA--A-GGAAAA .......((((((......(((((........))))).(((((.(.((((.....))))...).))))).............)--))))).......--.-...... ( -17.90) >DroPse_CAF1 52542 104 + 1 AAAGAGACAUUCAGCUGCAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAGUACAACUCCGAAUGAAUAACA--A-CAACAG ...(((((..(((((....(((((........))))).)))))...((((.....))))..)))))...............................--.-...... ( -19.30) >DroSim_CAF1 45147 96 + 1 AAAGAGACAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAA------U--GAAUGAACAGCA--A-GGAAAA .......((((((......(((((........))))).(((((.(.((((.....))))...).))))).......------)--))))).......--.-...... ( -17.90) >DroYak_CAF1 44816 105 + 1 AAAGAGAUAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAAUACAAAU--GAAUGAACAACAACAUGGAAAA .......((((((......(((((........))))).(((((.(.((((.....))))...).))))).............)--)))))................. ( -15.80) >DroAna_CAF1 41024 89 + 1 AAAGAGACACUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAAUACAAAU--GAAUGG---------------- ...(((((.........((((...(((.......))).))))....((((.....))))..))))).................--......---------------- ( -16.30) >DroPer_CAF1 43149 104 + 1 AAAGAGACAUUCAGCUGCAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAGUACAACUCCGAAUGAAUAACA--A-CAACAG ...(((((..(((((....(((((........))))).)))))...((((.....))))..)))))...............................--.-...... ( -19.30) >consensus AAAGAGACAUUCAACUACAGCAAUGCAUAUCAAUUGCCGCUGACAAUUUCAAAUAGAAAACGUCUCAGCAUGCCAAUACAAAU__GAAUGAACAACA__A_GGAAAA .......(((((.......(((((........))))).(((((.(.((((.....))))...).)))))................)))))................. (-16.32 = -16.35 + 0.03)


| Location | 12,689,124 – 12,689,226 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12689124 102 - 20766785 UUUUCC-U--UGCUGUUCAUUC--AUUUGUAUUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUGUCUCUUU ......-.--(((((..((...--...))...)))))....(((((((((.((...(((.....)))((((((((........))))))))))..)))))))))... ( -23.20) >DroPse_CAF1 52542 104 - 1 CUGUUG-U--UGUUAUUCAUUCGGAGUUGUACUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGCAGCUGAAUGUCUCUUU ......-.--.......(((((((((((((....))).)))..((((.((((.....)))).)))).((((((((........))))))))..)))))))....... ( -26.20) >DroSim_CAF1 45147 96 - 1 UUUUCC-U--UGCUGUUCAUUC--A------UUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUGUCUCUUU ......-.--.(((((.((...--.------.))))).)).(((((((((.((...(((.....)))((((((((........))))))))))..)))))))))... ( -22.20) >DroYak_CAF1 44816 105 - 1 UUUUCCAUGUUGUUGUUCAUUC--AUUUGUAUUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUAUCUCUUU .............((((((.((--(..(((....)))...)))((((.((((.....)))).)))).((((((((........))))))))...))))))....... ( -20.30) >DroAna_CAF1 41024 89 - 1 ----------------CCAUUC--AUUUGUAUUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAGUGUCUCUUU ----------------.(((((--(......(..(....)..)((((.((((.....)))).)))).((((((((........))))))))...))))))....... ( -21.00) >DroPer_CAF1 43149 104 - 1 CUGUUG-U--UGUUAUUCAUUCGGAGUUGUACUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGCAGCUGAAUGUCUCUUU ......-.--.......(((((((((((((....))).)))..((((.((((.....)))).)))).((((((((........))))))))..)))))))....... ( -26.20) >consensus UUUUCC_U__UGUUGUUCAUUC__AUUUGUAUUGGCAUGCUGAGACGUUUUCUAUUUGAAAUUGUCAGCGGCAAUUGAUAUGCAUUGCUGUAGUUGAAUGUCUCUUU .................................((((((((..((((.((((.....)))).)))).((((((((........)))))))))))...)))))..... (-20.86 = -20.45 + -0.41)

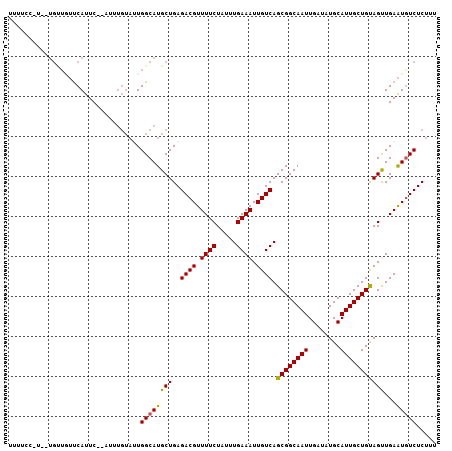
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:24 2006