| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,688,113 – 12,688,210 |
| Length | 97 |
| Max. P | 0.740514 |

| Location | 12,688,113 – 12,688,210 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -22.66 |
| Energy contribution | -22.27 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
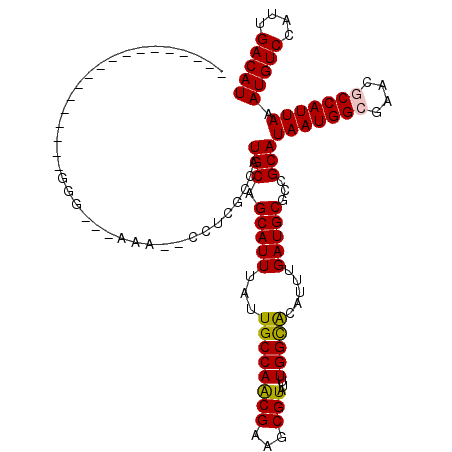
>2R_DroMel_CAF1 12688113 97 + 20766785 ------------------GGG---AAA--CCUCGCCAUGCAGCAUUUAUUGCCAGCGAAGCGUAUUUGGCACAUUUGAUGCGGCGCAUAAUGGGGAACGCCAUUAAAUGUCCAUUGACAU ------------------((.---...--))..((..(((.(((.....)))..)))..))((...(((.((((((((((((.(.(.....).)...)).)))))))))))))...)).. ( -29.10) >DroVir_CAF1 60853 91 + 1 -----------------------------CCUCGCCAUGCAGCAUUUAUUGCCAACGAAGCGUAUUUGGUGCAUUUGAUGCGCUGCAUAAUGGCGCACGCCAUUAAAUGUCCAUUGACAU -----------------------------........((((((.......(((((((...)))...))))((((...))))))))))(((((((....))))))).(((((....))))) ( -27.90) >DroGri_CAF1 46478 117 + 1 UAAAGGCAAAAAUAGAAAGGA---AAAACUCUAGCCAUGCAGCAUUUAUUGCCAACGAAGCGUAUUUGGUGCAUUUGAUGCGCUGCAUAAUGGCGCACGCCAUUAAAUGUCCAUUGACAU ....(((.....((((.....---.....))))))).((((((.......(((((((...)))...))))((((...))))))))))(((((((....))))))).(((((....))))) ( -31.80) >DroEre_CAF1 41870 97 + 1 ------------------GGG---AGA--ACUCGCCAUGCAGCAUUUAUUGCCAGCGAAGCGUAUUUGGCACAUUUGAUGCGGCGCAUAAUGGGGAACGCCAUUAAAUGUCCAUUGACAU ------------------(((---...--.)))((..(((.(((.....)))..)))..))((...(((.((((((((((((.(.(.....).)...)).)))))))))))))...)).. ( -25.60) >DroWil_CAF1 54819 100 + 1 ------------------GUGAAAAAA--CCUCACCAUGCAGCAUUUAUUGCCAACGAAGCGUAUUUGGCACAUUUGAUGCGGCGCAUAAUGGCAAACGCCAUUAAAUGUCCAUUGACAU ------------------((((.....--..))))...((.(((((...((((((((...)))...))))).....))))).))...(((((((....))))))).(((((....))))) ( -27.70) >DroMoj_CAF1 89883 91 + 1 -----------------------------CGUCGCCAUGCAGCAUUUAUUGCCAACGAAGCGUAUUUGGUGCAUUUGAUGCGCUGCAUAAUGGCGCACGCCAUUAAAUGUCCAUUGACAU -----------------------------((((((((((((((.......(((((((...)))...))))((((...))))))))))...))))).))).......(((((....))))) ( -29.00) >consensus __________________GGG___AAA__CCUCGCCAUGCAGCAUUUAUUGCCAACGAAGCGUAUUUGGCACAUUUGAUGCGCCGCAUAAUGGCGAACGCCAUUAAAUGUCCAUUGACAU .....................................(((.(((((...((((((((...)))...))))).....)))))...)))(((((((....))))))).(((((....))))) (-22.66 = -22.27 + -0.39)


| Location | 12,688,113 – 12,688,210 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
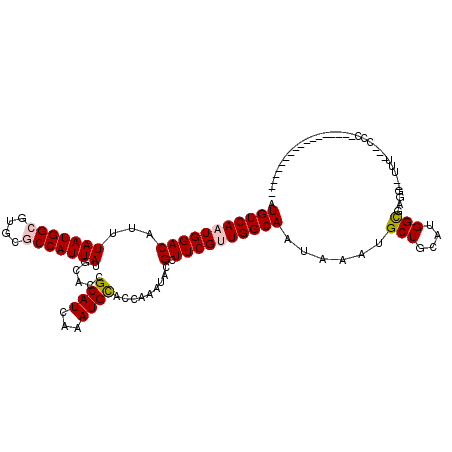
>2R_DroMel_CAF1 12688113 97 - 20766785 AUGUCAAUGGACAUUUAAUGGCGUUCCCCAUUAUGCGCCGCAUCAAAUGUGCCAAAUACGCUUCGCUGGCAAUAAAUGCUGCAUGGCGAGG--UUU---CCC------------------ .......(((((((((..((((((..........))))))....)))))).))).....((((((((((((........))).))))))))--)..---...------------------ ( -28.50) >DroVir_CAF1 60853 91 - 1 AUGUCAAUGGACAUUUAAUGGCGUGCGCCAUUAUGCAGCGCAUCAAAUGCACCAAAUACGCUUCGUUGGCAAUAAAUGCUGCAUGGCGAGG----------------------------- .(((((.........(((((((....)))))))((((((((((...)))).........(((.....))).......)))))))))))...----------------------------- ( -28.60) >DroGri_CAF1 46478 117 - 1 AUGUCAAUGGACAUUUAAUGGCGUGCGCCAUUAUGCAGCGCAUCAAAUGCACCAAAUACGCUUCGUUGGCAAUAAAUGCUGCAUGGCUAGAGUUUU---UCCUUUCUAUUUUUGCCUUUA (((((....))))).(((((((....)))))))((((((((((...)))).........(((.....))).......)))))).((((((((....---....))))).....))).... ( -33.00) >DroEre_CAF1 41870 97 - 1 AUGUCAAUGGACAUUUAAUGGCGUUCCCCAUUAUGCGCCGCAUCAAAUGUGCCAAAUACGCUUCGCUGGCAAUAAAUGCUGCAUGGCGAGU--UCU---CCC------------------ .......(((((((((..((((((..........))))))....)))))).)))....((((..((.((((.....))))))..))))...--...---...------------------ ( -26.00) >DroWil_CAF1 54819 100 - 1 AUGUCAAUGGACAUUUAAUGGCGUUUGCCAUUAUGCGCCGCAUCAAAUGUGCCAAAUACGCUUCGUUGGCAAUAAAUGCUGCAUGGUGAGG--UUUUUUCAC------------------ (((((....))))).(((((((....)))))))...((.((((......((((((..........))))))....)))).))...(((((.--....)))))------------------ ( -26.20) >DroMoj_CAF1 89883 91 - 1 AUGUCAAUGGACAUUUAAUGGCGUGCGCCAUUAUGCAGCGCAUCAAAUGCACCAAAUACGCUUCGUUGGCAAUAAAUGCUGCAUGGCGACG----------------------------- (((((....))))).......(((.(((((...((((((((((...)))).........(((.....))).......))))))))))))))----------------------------- ( -28.90) >consensus AUGUCAAUGGACAUUUAAUGGCGUGCGCCAUUAUGCACCGCAUCAAAUGCACCAAAUACGCUUCGUUGGCAAUAAAUGCUGCAUGGCGAGG__UUU___CCC__________________ .(((((((((((...(((((((....)))))))......((((...)))).........).))))))))))......(((....)))................................. (-19.95 = -20.23 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:20 2006