
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,652,439 – 12,652,545 |
| Length | 106 |
| Max. P | 0.647093 |

| Location | 12,652,439 – 12,652,545 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.90 |
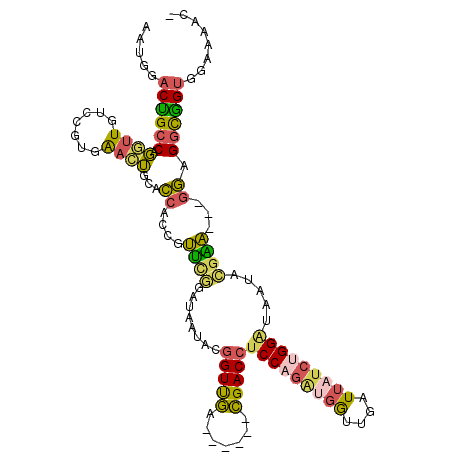
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
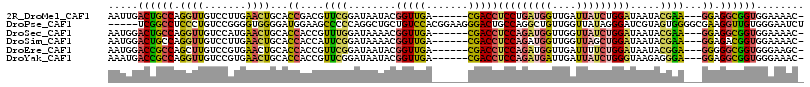
>2R_DroMel_CAF1 12652439 106 - 20766785 AAUUGACUGCCAGGUUGUCCUUGAACUGCACCGACGUUCGGAUAAUACGGUUGA------CGACCUCCUGAUGGUUGAUUAUCUGGAUAAUACGAA---GGAGGCGGUGGAAAAC- ..((.((((((.(((((((....(((((..(((.....)))......)))))))------)))))((((..((....(((((....))))).)).)---))))))))).))....- ( -31.90) >DroPse_CAF1 13056 111 - 1 -----UCGGCCUCCCUGUCCGGGGUGGGGAUGGAAGCCCCCAGGCUGCUGUCCACGGAAGGGACUGCCAGGCUGUUGGUUAUAGGGAUCGUAGUGGGGCGAAGGUUGUGGGAAUCU -----.(((((((((((((((((((..........)))))(((.((((.((((.(....))))).).))).)))..))..)))))))((((......)))).)))))......... ( -44.50) >DroSec_CAF1 10595 106 - 1 AAUGGACUGCCAGGUUGUCCAUGAACUGCACCACCGUUUGGAUAAAACGGUUGA------CGACCUCCAGAUGGUUGGUUAUCUGGAUAAUACGAA---GGAGGCGGUGGAAAAC- .....((((((..((((((((.((.....(((((((((((((......((((..------.))))))))))))).))))..)))))))))).(...---.).)))))).......- ( -36.60) >DroSim_CAF1 12162 106 - 1 AAUGGACUGCCAGGUUGUCCUUGAACUGCACCACCAUUCGGAUAAAACGGUUGA------CGACCUCCAGAUGGUUGGUUAGCUGGAUAAUACGAA---GGAGACGGUGGAAAAC- ...((((.((...)).))))..........(((((.((((.......(((((((------(((((.......)))).)))))))).......))))---(....)))))).....- ( -33.64) >DroEre_CAF1 9479 106 - 1 AAUGGACCGCCAGCUUGUCCGUGAACUGCACCACCGUUCGGAUAAUACGGUUGA------CGACCUCCAGAUGGUUGAUUUUCUGGAUAAUACGGA---GGGGGCGGUGGGAAGC- ......(((((..(((.((((((.......(.(((((.........))))).).------.....((((((..........))))))...))))))---.)))..))))).....- ( -33.30) >DroYak_CAF1 11464 106 - 1 AAAUGACCGCCAGGUUGUCCGUGAACUGCACCACCGUUCGGAUAAUACGGUUGA------CGACCUCCAGAUGAUUGAUUAUCUGGGUAAGAGGGA---GGAGGCGGUGGGAAAC- ...(.((((((((((((((.(((.....))).(((((.........))))).))------))))))((((((((....))))))))..........---...)))))).).....- ( -38.80) >consensus AAUGGACUGCCAGGUUGUCCGUGAACUGCACCACCGUUCGGAUAAUACGGUUGA______CGACCUCCAGAUGGUUGAUUAUCUGGAUAAUACGAA___GGAGGCGGUGGAAAAC_ .....((((((.((((.......))))...((....((((........(((((.......)))))(((((((((....))))))))).....))))...)).))))))........ (-21.80 = -21.20 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:01 2006