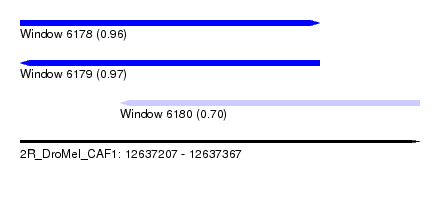
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,637,207 – 12,637,367 |
| Length | 160 |
| Max. P | 0.973483 |

| Location | 12,637,207 – 12,637,327 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12637207 120 + 20766785 UAUUUAGUUUCCGGUUUUAACUUGCGCCAUAAACCGGGCGUUAUGUGUCCGCUUUUAUGGCUUCUAGCGACGUCCCGUCUUCAUUGUUUCUUAAUAAUCCGACGAGCGUCUUCGGAGUGC ........((((((...........((((((((.((((((.....))))))..)))))))).......(((((..((((...((((((....))))))..)))).))))).))))))... ( -36.60) >DroSec_CAF1 29903 120 + 1 UAUUUUGUUUCCGGUUUUAACUUGCGCCAUAAAGCGGGCGUUAUGUGUCCGCUUUUAUGGCUUCUAGCGACGUCCCGUCUUCAUUGUUUCUUAAUAAUCCGACGAGCGUCUUCGGAGUGC ........((((((...........(((((((((((((((.....))))))).)))))))).......(((((..((((...((((((....))))))..)))).))))).))))))... ( -41.30) >DroEre_CAF1 30805 120 + 1 UAUUUAGUUUCCGGUUUUAACUUGCGCCAUAAACCGGGCGUUAUGUGUCCGCUUUUAUGGCUUCUAGCGACGUCCCGUCUUCAUUGUUUCUUAAUAAUCCGACGAGCGUCUUCGGAGUGC ........((((((...........((((((((.((((((.....))))))..)))))))).......(((((..((((...((((((....))))))..)))).))))).))))))... ( -36.60) >consensus UAUUUAGUUUCCGGUUUUAACUUGCGCCAUAAACCGGGCGUUAUGUGUCCGCUUUUAUGGCUUCUAGCGACGUCCCGUCUUCAUUGUUUCUUAAUAAUCCGACGAGCGUCUUCGGAGUGC ........((((((...........((((((((.((((((.....))))))..)))))))).......(((((..((((...((((((....))))))..)))).))))).))))))... (-36.00 = -36.00 + 0.00)

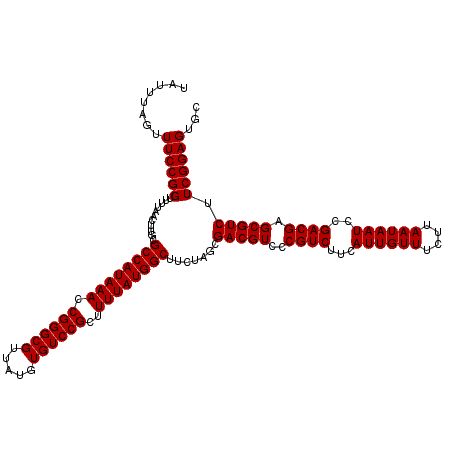
| Location | 12,637,207 – 12,637,327 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -34.53 |
| Energy contribution | -34.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12637207 120 - 20766785 GCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAACGCCCGGUUUAUGGCGCAAGUUAAAACCGGAAACUAAAUA ....((((..((((((((((...(((((.......))))).)))))).))))((.....))........))))..........(((((((.((((....))))))))))).......... ( -36.10) >DroSec_CAF1 29903 120 - 1 GCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAACGCCCGCUUUAUGGCGCAAGUUAAAACCGGAAACAAAAUA ....((((..((((((((((...(((((.......))))).)))))).))))(((....((((((((.((((.(.......).))))))))))))...)))......))))......... ( -38.60) >DroEre_CAF1 30805 120 - 1 GCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAACGCCCGGUUUAUGGCGCAAGUUAAAACCGGAAACUAAAUA ....((((..((((((((((...(((((.......))))).)))))).))))((.....))........))))..........(((((((.((((....))))))))))).......... ( -36.10) >consensus GCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAACGCCCGGUUUAUGGCGCAAGUUAAAACCGGAAACUAAAUA ....((((..((((((((((...(((((.......))))).)))))).))))(((....((((((((.(((.........)))....))))))))...)))......))))......... (-34.53 = -34.53 + 0.00)


| Location | 12,637,247 – 12,637,367 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -34.00 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12637247 120 - 20766785 CAGUGUCUGGCCCAUAAACAUCCUUAAUGCGAGGACGUGUGCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAA ..(((((((...........(((((.....))))).(((.((..((....((((((((((...(((((.......))))).)))))).))))....)).))))).....))))))).... ( -34.00) >DroSec_CAF1 29943 120 - 1 CAGUGUCUGGCCCAUAAACAUCCUUAAUGCGAGGACGUGUGCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAA ..(((((((...........(((((.....))))).(((.((..((....((((((((((...(((((.......))))).)))))).))))....)).))))).....))))))).... ( -34.00) >DroEre_CAF1 30845 120 - 1 CAGUGUCUGGCCCAUAAACAUCCUUAAUGCGAGGACGUGUGCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAA ..(((((((...........(((((.....))))).(((.((..((....((((((((((...(((((.......))))).)))))).))))....)).))))).....))))))).... ( -34.00) >consensus CAGUGUCUGGCCCAUAAACAUCCUUAAUGCGAGGACGUGUGCACUCCGAAGACGCUCGUCGGAUUAUUAAGAAACAAUGAAGACGGGACGUCGCUAGAAGCCAUAAAAGCGGACACAUAA ..(((((((...........(((((.....))))).(((.((..((....((((((((((...(((((.......))))).)))))).))))....)).))))).....))))))).... (-34.00 = -34.00 + 0.00)

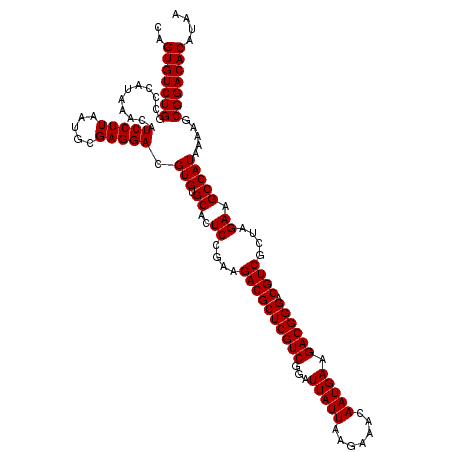
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:56 2006