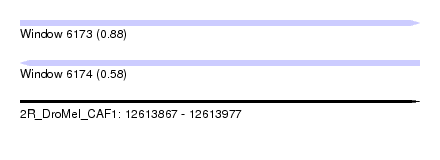
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,613,867 – 12,613,977 |
| Length | 110 |
| Max. P | 0.879897 |

| Location | 12,613,867 – 12,613,977 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -10.12 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
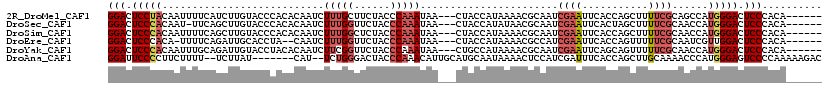
>2R_DroMel_CAF1 12613867 110 + 20766785 GGACUCCUACAAUUUUCAUCUUGUACCCACACAAUCUUUGCUUCUACCCAAAUAA---CUACCAUAAAACGCAAUCGAAUUCACCAGCUUUUCGCAGCCAUGGGACUCCCACA------ (((.(((((.....(((...((((......))))...((((..............---............))))..))).......(((......)))..))))).)))....------ ( -12.37) >DroSec_CAF1 6761 109 + 1 GGACUCCCACAAU-UUCAGCUUGUACCCACACAAUCUUUGGUUCUACCCAAAUAA---CUACCAUAUAACGCAAUCGAAUUCACUAGCUUUUCGCAACCAUGGGACUCCCACA------ (((.(((((....-....((((((......))))..(((((......)))))...---............))..............((.....)).....))))).)))....------ ( -18.20) >DroSim_CAF1 6775 110 + 1 GGACUCCCACAAUUUUCAGCUUGUACCCACACAAUCUUUGGCUCUACCCAAAUAA---CUACCAUAAAACGCAAUCGAAUUCACCAGCUUUUCGCAACCAUGGGACUCCCACA------ (((.(((((.........((((((......))))..(((((......)))))...---............))..............((.....)).....))))).)))....------ ( -17.90) >DroEre_CAF1 7029 107 + 1 GGACUCCCACA-UUUUCAGAUUGCACCUA--CAAUCUUUGGUUCUACCCAAAUAA---CUACCAUAAAACGCCAUCGAAUUCACCAGUUUUUCGCAAUCGUUGGACUCCCACA------ (((.(((.((.-......((((((.....--.....(((((......)))))...---.......(((((................)))))..)))))))).))).)))....------ ( -17.20) >DroYak_CAF1 6874 110 + 1 GGACUCCCACAAUUUGCAGAUUGUACCUACACAAUCUUCGGUUCUACCCAAAUAA---CUGCCAUAAAACGCAAUCGAAUUCAGCAGUUUUUCGCAACCAUGGGACUCCCACA------ (((.(((((....(((((((((((......)))))))..((......))....((---((((.......((....))......))))))....))))...))))).)))....------ ( -26.22) >DroAna_CAF1 7176 108 + 1 GGAUUCCCCUUCUUUU--UCUUAU-------CAU--UCUGGGACUACCCAAACAUUGCAUGCAAUAAAACUCCAUCGAUUUCACCAGCUUGCAAAACCCAUGGGAGUCCCCAAAAAGAC ((((((((........--......-------...--..((((....))))....(((((.((........................)).))))).......)))))))).......... ( -23.76) >consensus GGACUCCCACAAUUUUCAGCUUGUACCCACACAAUCUUUGGUUCUACCCAAAUAA___CUACCAUAAAACGCAAUCGAAUUCACCAGCUUUUCGCAACCAUGGGACUCCCACA______ (((.(((((...........................(((((......))))).......................((((...........))))......))))).))).......... (-10.12 = -11.07 + 0.95)
| Location | 12,613,867 – 12,613,977 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -16.95 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
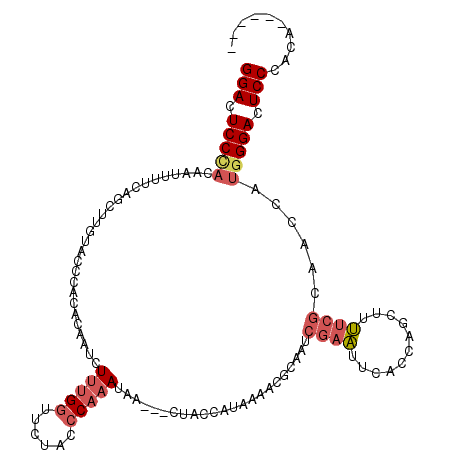
>2R_DroMel_CAF1 12613867 110 - 20766785 ------UGUGGGAGUCCCAUGGCUGCGAAAAGCUGGUGAAUUCGAUUGCGUUUUAUGGUAG---UUAUUUGGGUAGAAGCAAAGAUUGUGUGGGUACAAGAUGAAAAUUGUAGGAGUCC ------....(((.(((....((..((((...(....)..))))...))....((..((..---((((((..(((...(((.......)))...))).))))))..))..))))).))) ( -21.50) >DroSec_CAF1 6761 109 - 1 ------UGUGGGAGUCCCAUGGUUGCGAAAAGCUAGUGAAUUCGAUUGCGUUAUAUGGUAG---UUAUUUGGGUAGAACCAAAGAUUGUGUGGGUACAAGCUGAA-AUUGUGGGAGUCC ------....(((.(((((..(((......((((.(((....(((((((........))))---)).(((((......)))))......)....))).))))..)-))..))))).))) ( -32.20) >DroSim_CAF1 6775 110 - 1 ------UGUGGGAGUCCCAUGGUUGCGAAAAGCUGGUGAAUUCGAUUGCGUUUUAUGGUAG---UUAUUUGGGUAGAGCCAAAGAUUGUGUGGGUACAAGCUGAAAAUUGUGGGAGUCC ------....(((.(((((..(((......((((.(((....(((((((........))))---)).(((((......)))))......)....))).))))...)))..))))).))) ( -31.60) >DroEre_CAF1 7029 107 - 1 ------UGUGGGAGUCCAACGAUUGCGAAAAACUGGUGAAUUCGAUGGCGUUUUAUGGUAG---UUAUUUGGGUAGAACCAAAGAUUG--UAGGUGCAAUCUGAAAA-UGUGGGAGUCC ------....(((.(((..((((..(.(.....).)..)..)))...(((((((.......---....((((......))))((((((--(....))))))).))))-))).))).))) ( -26.20) >DroYak_CAF1 6874 110 - 1 ------UGUGGGAGUCCCAUGGUUGCGAAAAACUGCUGAAUUCGAUUGCGUUUUAUGGCAG---UUAUUUGGGUAGAACCGAAGAUUGUGUAGGUACAAUCUGCAAAUUGUGGGAGUCC ------....(((.(((((..(((((....((((((((((..((....))..)).))))))---))..((((......))))((((((((....)))))))))).)))..))))).))) ( -40.50) >DroAna_CAF1 7176 108 - 1 GUCUUUUUGGGGACUCCCAUGGGUUUUGCAAGCUGGUGAAAUCGAUGGAGUUUUAUUGCAUGCAAUGUUUGGGUAGUCCCAGA--AUG-------AUAAGA--AAAAGAAGGGGAAUCC (((.(((((((.(((((((......(((((.((.((((((((.......)))))))))).)))))....)))).)))))))))--).)-------))....--................ ( -31.20) >consensus ______UGUGGGAGUCCCAUGGUUGCGAAAAGCUGGUGAAUUCGAUUGCGUUUUAUGGUAG___UUAUUUGGGUAGAACCAAAGAUUGUGUGGGUACAAGCUGAAAAUUGUGGGAGUCC ..........(((.((((((..((((.((((((...((....)).....))))).).))))......(((((......)))))..........................)))))).))) (-16.95 = -16.57 + -0.38)

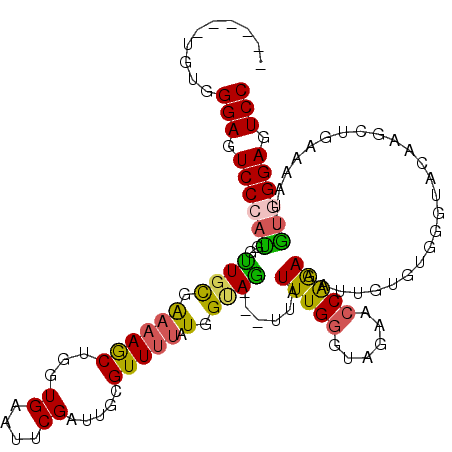
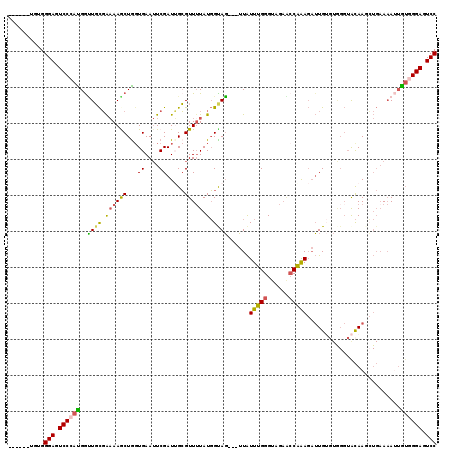
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:50 2006