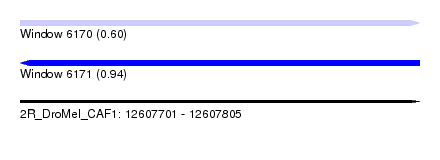
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,607,701 – 12,607,805 |
| Length | 104 |
| Max. P | 0.939936 |

| Location | 12,607,701 – 12,607,805 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -20.21 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12607701 104 + 20766785 AGAUGUGCGAAAAUUUCCAGCUGGGCGACACCAAAGAGGGCAUUGCUAGCUUCUUCGAGAAGCGCCCACCCAACUGGAAGCACCAGUAACCG-----CAACAUACAAUA ..(((((((.....((((((.((((.(....).....((((.......(((((.....))))))))).)))).))))))((....))...))-----).))))...... ( -31.11) >DroSec_CAF1 776 104 + 1 AGAUGUGCGAAAAUUUCCAGCUGGGUGACACCAAAGAGGGCAUUGCUAGCUUCUUCGAGAAGCGUCCACCCAACUGGAAGCACCAGUAACCG-----CAACCUACAAUG .....((((.....((((((.((((((((.((.....)).........(((((.....))))))).)))))).))))))((....))...))-----)).......... ( -33.30) >DroSim_CAF1 761 104 + 1 AGAUGUGCGAAAAUUUCCAGCUGGGCGACACCAAAGAGGGCAUUUCUAGCUUCUUCGAGAAGCGUCCACCCAACUGGAAGCACCAGUAACCG-----CAACCUGCAAUG ....((((......((((((.((((.(((.((.....)).........(((((.....))))))))..)))).))))))))))........(-----(.....)).... ( -31.20) >DroEre_CAF1 827 103 + 1 AGAUGUGCGAGAACUUCCAGCUGGGCGACACCAAAGAAGGCAUUGCUAGCUUCUUCGAGAAGCGCCCACCCAACUGGAAGCACAACUAA-CA-----UAAUCUAUUAUG ((.(((((......((((((.(((((((..((......))..))))..(((((.....)))))......))).))))))))))).))..-((-----(((....))))) ( -30.70) >DroYak_CAF1 827 104 + 1 AGAUGUGCGAGAACUUUCAGCUGGGCGACACCAAAGAGGGCAUUGCUAGCUUCUUUGAGAAGCGUCCACCCAACUGGAAGCACAACUAAUCG-----CAACGUACGAUG ..((((((((...(((((((.((((.(((.((.....)).........(((((.....))))))))..)))).))))))).........)))-----).))))...... ( -32.00) >DroPer_CAF1 6001 109 + 1 AAAUGGAAGAGAACCUGCAAUUGAGCGACUGCCAGGAAGGCAUCGCUAGCUUCAUAGAGAAGCGUGCACCCCACUGGAAGCACGAGUAAUACCUGUAUAUCCUAUCCUU ....(((((.((...((((....(((((.((((.....))))))))).(((((.....)))))((((..((....))..))))..........))))..)))).))).. ( -31.90) >consensus AGAUGUGCGAAAACUUCCAGCUGGGCGACACCAAAGAGGGCAUUGCUAGCUUCUUCGAGAAGCGUCCACCCAACUGGAAGCACCAGUAACCG_____CAACCUACAAUG ....((((......((((((.(((((((..((......))..))))..(((((.....)))))......))).)))))))))).......................... (-20.21 = -21.52 + 1.31)

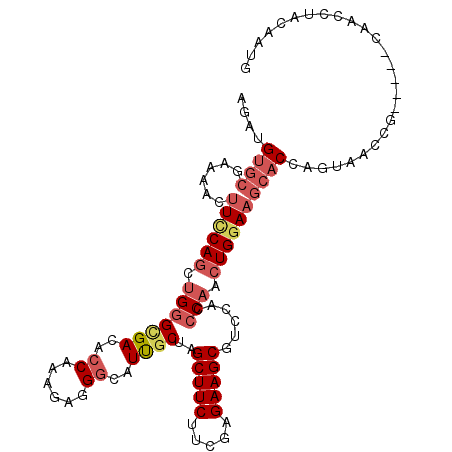
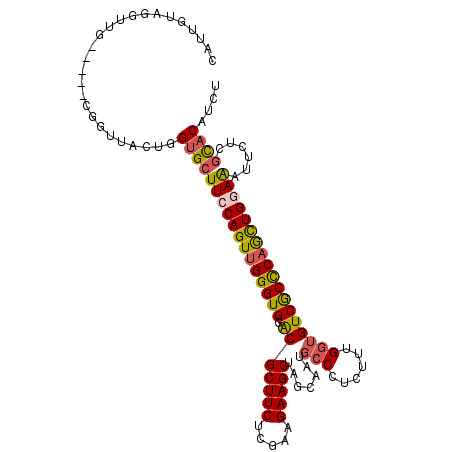
| Location | 12,607,701 – 12,607,805 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -31.33 |
| Energy contribution | -31.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12607701 104 - 20766785 UAUUGUAUGUUG-----CGGUUACUGGUGCUUCCAGUUGGGUGGGCGCUUCUCGAAGAAGCUAGCAAUGCCCUCUUUGGUGUCGCCCAGCUGGAAAUUUUCGCACAUCU ......((((.(-----((((.......))(((((((((((((((((((((.....)))))..))...(((......))).)))))))))))))).....))))))).. ( -37.80) >DroSec_CAF1 776 104 - 1 CAUUGUAGGUUG-----CGGUUACUGGUGCUUCCAGUUGGGUGGACGCUUCUCGAAGAAGCUAGCAAUGCCCUCUUUGGUGUCACCCAGCUGGAAAUUUUCGCACAUCU ......((((((-----((((.......))(((((((((((((.((((....((((((.((.......))..))))))))))))))))))))))).....)))).)))) ( -38.60) >DroSim_CAF1 761 104 - 1 CAUUGCAGGUUG-----CGGUUACUGGUGCUUCCAGUUGGGUGGACGCUUCUCGAAGAAGCUAGAAAUGCCCUCUUUGGUGUCGCCCAGCUGGAAAUUUUCGCACAUCU .(((((.....)-----)))).....(((((((((((((((((.((((....((((((.((.......))..)))))))))))))))))))))))......)))).... ( -40.00) >DroEre_CAF1 827 103 - 1 CAUAAUAGAUUA-----UG-UUAGUUGUGCUUCCAGUUGGGUGGGCGCUUCUCGAAGAAGCUAGCAAUGCCUUCUUUGGUGUCGCCCAGCUGGAAGUUCUCGCACAUCU ......((((..-----((-(.((....((((((((((((((((.(((....((((((((.((....)).))))))))))))))))))))))))))).)).))).)))) ( -43.30) >DroYak_CAF1 827 104 - 1 CAUCGUACGUUG-----CGAUUAGUUGUGCUUCCAGUUGGGUGGACGCUUCUCAAAGAAGCUAGCAAUGCCCUCUUUGGUGUCGCCCAGCUGAAAGUUCUCGCACAUCU .(((((.....)-----)))).((.(((((...((((((((((.((((....((((((.((.......))..))))))))))))))))))))...(....)))))).)) ( -36.30) >DroPer_CAF1 6001 109 - 1 AAGGAUAGGAUAUACAGGUAUUACUCGUGCUUCCAGUGGGGUGCACGCUUCUCUAUGAAGCUAGCGAUGCCUUCCUGGCAGUCGCUCAAUUGCAGGUUCUCUUCCAUUU ..(((..(((.....((((((..((.((((.(((....))).))))(((((.....))))).))..)))))).((((.((((......)))))))).)))..))).... ( -37.30) >consensus CAUUGUAGGUUG_____CGGUUACUGGUGCUUCCAGUUGGGUGGACGCUUCUCGAAGAAGCUAGCAAUGCCCUCUUUGGUGUCGCCCAGCUGGAAAUUCUCGCACAUCU ..........................(((((((((((((((((.(((((((.....))))).......(((......))))))))))))))))))......)))).... (-31.33 = -31.45 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:47 2006