
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,595,542 – 12,595,656 |
| Length | 114 |
| Max. P | 0.508997 |

| Location | 12,595,542 – 12,595,656 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
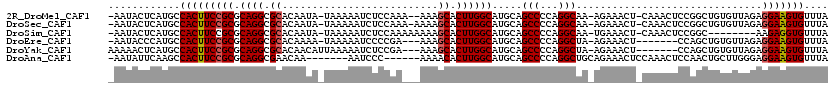
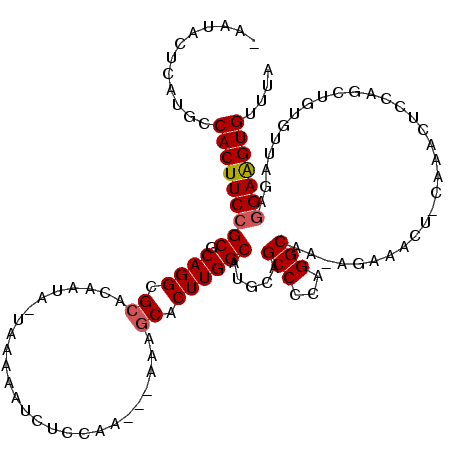
>2R_DroMel_CAF1 12595542 114 - 20766785 -AAUACUCAUGCCACUUCCGCGCAGGCGCACAAUA-UAAAAAUCUCCAAA--AAAGCACUUGGCAUGCAGCCCCAGGCAA-AGAAACU-CAAACUCCGGCUGUGUUAGAGGAAGUGUUUA -...........(((((((((.((((.((......-..............--...)).))))))((((((((..((....-.......-....))..))))))))....))))))).... ( -28.56) >DroSec_CAF1 23115 115 - 1 -AAUACUCAUGCCACUUCCGCGCAGGCGCACAAUA-UAAAAAUCUCCAAA-AAAAGCACUUGGCAUGCAGCCCCAGGCAA-AGAAACU-CAAACUCCGGCUGUGUUAGAGGAAGUGUUUA -...........(((((((((.((((.((......-..............-....)).))))))((((((((..((....-.......-....))..))))))))....))))))).... ( -28.52) >DroSim_CAF1 20723 108 - 1 -AAUACUCAUGCCACUUCCGCGCAGGCGCACAAUA-UAAAAAUCUCCAAAAAAAAGCACUUGGCAUGCAGCCCCAGGCAA-UGAAACU-CAAACUCCGGC--------AAGAGGUGUUUA -....(((.((((......(((....)))......-...................((....(((.....)))....))..-.......-........)))--------).)))....... ( -22.70) >DroEre_CAF1 20493 107 - 1 -AAUACCCAUGCCACUUCCGCGCAGGCGCACAAAA-UAAAAAUCCCCGA---AAAGCACUUGGCAUGCAGCCCCAGGCUA-AGAAACU-------CCAGCUGUGUUAGAGGAAGUGUUUA -...........((((((((((....)))......-.............---..(((((..(((.....)))...((((.-.......-------..)))))))))...))))))).... ( -25.60) >DroYak_CAF1 21303 109 - 1 AAAAACUCAUGCCACUUCCGCGCAGGCGCACAACAUUAAAAAUCUCCGA---AAAGCACUUGGCAUGCAGCCCCAGGCUA-AGAAACU-------CCAGCUGUGUUAGAGGAAGUGUUUA ............((((((((((....)))....................---..(((((..(((.....)))...((((.-.......-------..)))))))))...))))))).... ( -25.60) >DroAna_CAF1 20277 106 - 1 -AAUAUUCAAGCCACUUCCGCGCAGGCGAACAA-------AAUCCC------AAAACACUUGGCAUGCAGCCCCAGGCUGCAGAAACUCCAAACUCCAACUGCUUGGGAGGAAGUGUUUA -...........(((((((.(.((((((.....-------....((------((.....))))..(((((((...)))))))..................)))))).).))))))).... ( -34.30) >consensus _AAUACUCAUGCCACUUCCGCGCAGGCGCACAAUA_UAAAAAUCUCCAA___AAAGCACUUGGCAUGCAGCCCCAGGCAA_AGAAACU_CAAACUCCAGCUGUGUUAGAGGAAGUGUUUA ............(((((((((.((((.((..........................)).)))))).....(((...)))...............................))))))).... (-19.31 = -19.50 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:42 2006