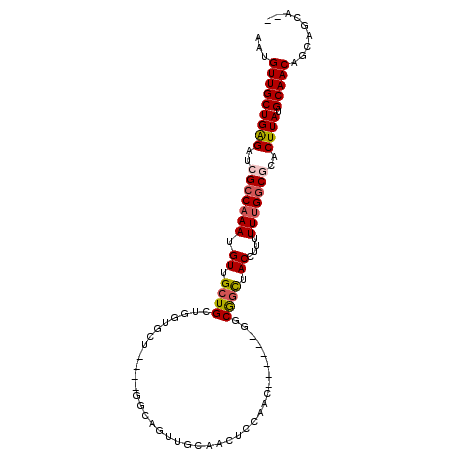
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,591,526 – 12,591,626 |
| Length | 100 |
| Max. P | 0.991630 |

| Location | 12,591,526 – 12,591,626 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.99 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12591526 100 + 20766785 ACUGCUGCUGUUGCAUAAGUGUGCCAAAAAAGGUGGCCGCC------GUUGGAGUUGCAACUGCGAAACAGCACCGCCAGCAACAUUUGGCGGUCCCAGCAACAUG ..(((((((((((((...(((.((((.......))))))).------((((......)))))))..))))))((((((((......))))))))...))))..... ( -36.40) >DroVir_CAF1 22598 92 + 1 --UG-UACAGUUGCAUAAGUGCGCCAAAAAAGGUAGCUGUAAAUGCAGUUGCAGUUGCAGUUGC-----------CGCAUCAACAUUGGCCGAUCUCAGCAACAUU --..-....(((((.....((.(((((....(((((((((((.(((....))).))))))))))-----------)(......).)))))))......)))))... ( -32.50) >DroPse_CAF1 17141 94 + 1 --UGAAUAAGUUGCAUAAGUGCGCCAAAAAAGGUAACCGCC------AUCGGAGUUGCAUGUGAC----AACGGCAGCAGCAACAUUUGGCGAGCUCAGCAACAUU --.......(((((...(((.(((((((....((..(((..------..))).(((((.(((...----.)))))))).))....))))))).)))..)))))... ( -30.90) >DroSec_CAF1 16506 100 + 1 ACUGCUGAUGUUGCAUAAGUGUGCCAAAAAAGGUGGCCGCC------GUUGGAGUUGCAACUGACAAAAAGCACCACCAGCAACAUUUGGCGACCCCAGCAACAUG ..(((((..(((((.(((((((((((.......))))....------(((((.(.(((............))).).))))).))))))))))))..)))))..... ( -33.00) >DroYak_CAF1 16574 100 + 1 ACUGCCGCUGUUGCAUAAGUGCGCCAAAAAAGGUAGCCGCA------GUUGGAGUUGCAACGCCGAAACUGCACCACCAGCAACAUUUGGCGAUCCCAGCAACAUG (((((.(((...(((....)))(((......)))))).)))------))....(((((..(((((((..(((.......)))...)))))))......)))))... ( -31.20) >DroPer_CAF1 17210 94 + 1 --UGAAUAAGUUGCAUAAGUGCGCCAAAAAAGGUAACCGCC------AUCGGAGUUGCAUGUGAC----AACGGCAGCAGCAACAUUUGGCGAGCUCAGCAACAUU --.......(((((...(((.(((((((....((..(((..------..))).(((((.(((...----.)))))))).))....))))))).)))..)))))... ( -30.90) >consensus __UGCUGAAGUUGCAUAAGUGCGCCAAAAAAGGUAGCCGCC______GUUGGAGUUGCAACUGAC____AGCACCACCAGCAACAUUUGGCGAUCCCAGCAACAUG .........(((((.......(((((((........(((..........))).(((((.....................))))).)))))))......)))))... (-17.90 = -17.99 + 0.09)

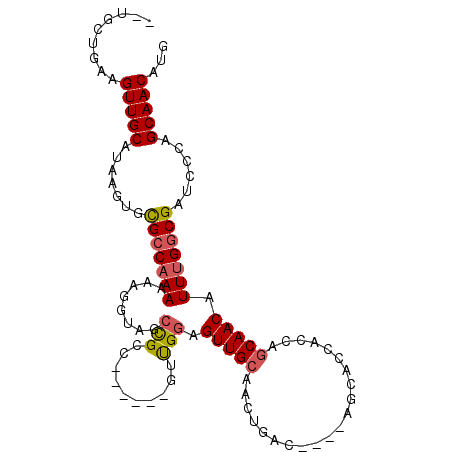
| Location | 12,591,526 – 12,591,626 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12591526 100 - 20766785 CAUGUUGCUGGGACCGCCAAAUGUUGCUGGCGGUGCUGUUUCGCAGUUGCAACUCCAAC------GGCGGCCACCUUUUUUGGCACACUUAUGCAACAGCAGCAGU ..(((((((((.(((((((........))))))).)(((..(((.((((......))))------.)))((((.......))))........))).)))))))).. ( -38.80) >DroVir_CAF1 22598 92 - 1 AAUGUUGCUGAGAUCGGCCAAUGUUGAUGCG-----------GCAACUGCAACUGCAACUGCAUUUACAGCUACCUUUUUUGGCGCACUUAUGCAACUGUA-CA-- ...(((((((..((((((....)))))).))-----------)))))......((((..(((((.....((((.......))))......)))))..))))-..-- ( -25.50) >DroPse_CAF1 17141 94 - 1 AAUGUUGCUGAGCUCGCCAAAUGUUGCUGCUGCCGUU----GUCACAUGCAACUCCGAU------GGCGGUUACCUUUUUUGGCGCACUUAUGCAACUUAUUCA-- ...(((((((((..(((((((.......(((((((((----(.............))))------)))))).......)))))))..)))).))))).......-- ( -34.06) >DroSec_CAF1 16506 100 - 1 CAUGUUGCUGGGGUCGCCAAAUGUUGCUGGUGGUGCUUUUUGUCAGUUGCAACUCCAAC------GGCGGCCACCUUUUUUGGCACACUUAUGCAACAUCAGCAGU .(((((((((((((.((((((.......((((((......((((.((((......))))------))))))))))...)))))))).)))).)))))))....... ( -32.90) >DroYak_CAF1 16574 100 - 1 CAUGUUGCUGGGAUCGCCAAAUGUUGCUGGUGGUGCAGUUUCGGCGUUGCAACUCCAAC------UGCGGCUACCUUUUUUGGCGCACUUAUGCAACAGCGGCAGU ..((((((((.....((((((.......((((((((((((..((.((....)).)))))------))).))))))...))))))(((....)))..)))))))).. ( -39.00) >DroPer_CAF1 17210 94 - 1 AAUGUUGCUGAGCUCGCCAAAUGUUGCUGCUGCCGUU----GUCACAUGCAACUCCGAU------GGCGGUUACCUUUUUUGGCGCACUUAUGCAACUUAUUCA-- ...(((((((((..(((((((.......(((((((((----(.............))))------)))))).......)))))))..)))).))))).......-- ( -34.06) >consensus AAUGUUGCUGAGAUCGCCAAAUGUUGCUGCUGGUGCU____GGCAGUUGCAACUCCAAC______GGCGGCUACCUUUUUUGGCGCACUUAUGCAACAGCAGCA__ ...(((((((((..(((((((.((.((((......................................)))).))....)))))))..)))).)))))......... (-17.75 = -17.98 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:40 2006