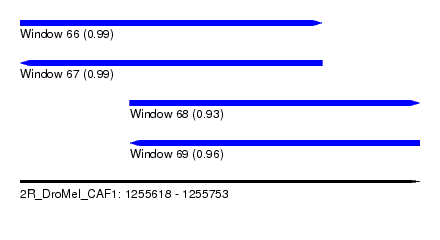
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,255,618 – 1,255,753 |
| Length | 135 |
| Max. P | 0.987314 |

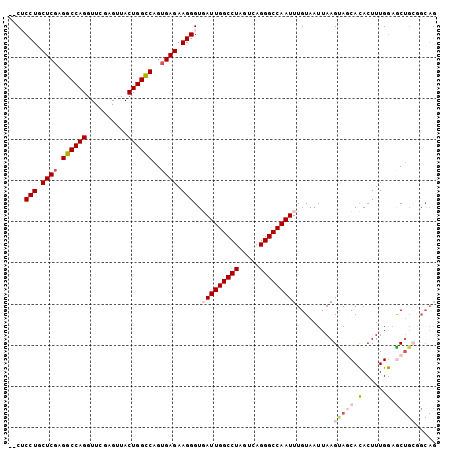
| Location | 1,255,618 – 1,255,720 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1255618 102 + 20766785 CUGCAUCAGCUCCAAAGUGUGCUACUUAAUUACAAAUUGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGAG-- ..((((..(((....))))))).............(((((((.......)))))))...(((.(((...((((((..........))))))..))).)))..-- ( -29.90) >DroSec_CAF1 7335 104 + 1 CUGCCGCAGCUCCAAAGUGUGCUGCUUAAUUACAAAUUGGCCCUGACUAGGCCAAUCACCCUUCUCCCUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGUGAA .....(((((.(......).)))))...........((((((.......))))))(((((...(((...((((((..........))))))..)))..))))). ( -36.60) >DroSim_CAF1 13161 104 + 1 CUGCCGCAGCUCCAAAGUGUGCUGCUUAAUUACAAAUUGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGUGGA .....(((((.(......).)))))...........((((((.......))))))(((((...(((...((((((..........))))))..)))..))))). ( -35.70) >DroEre_CAF1 11548 94 + 1 --------GUACCAAAGUGUGGUACUUAAUUACAAAUUGGCCCUGAGUAGGCCAAUAACCCUUCUCACUGGCCAGUAACUUGAACCUGGCCUCGAGCAGGAG-- --------((((((.....))))))..........(((((((.......)))))))...(((.(((...((((((..........))))))..))).)))..-- ( -34.00) >DroYak_CAF1 10828 102 + 1 CUGCCGCAGUUUCAAAGUAUGGUACUUAAUUACAAAUUGGCCCUGACUAGGCCAAUAACCCUUCUCACUGACCAGUAUCUCGAACCUGGCCUCGAGAAGGAG-- .(((((.(.(.....).).)))))...........(((((((.......)))))))...(((((((...(.((((..........)))).)..)))))))..-- ( -26.00) >consensus CUGCCGCAGCUCCAAAGUGUGCUACUUAAUUACAAAUUGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGAG__ ..............((((.....))))........(((((((.......)))))))...(((.(((...((((((..........))))))..))).))).... (-25.82 = -25.78 + -0.04)

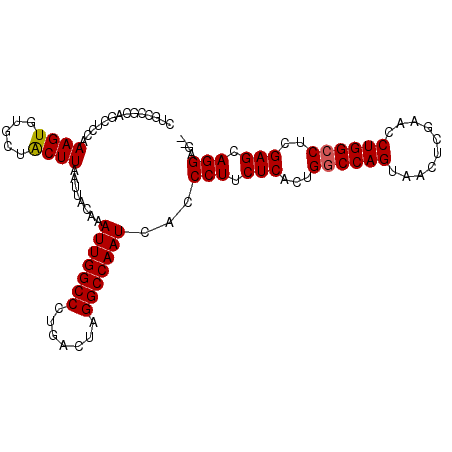
| Location | 1,255,618 – 1,255,720 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -30.40 |
| Energy contribution | -31.96 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1255618 102 - 20766785 --CUCCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCAAUUUGUAAUUAAGUAGCACACUUUGGAGCUGAUGCAG --...((((((..((((((..........))))))(((..((((.(((..((((((.....))))))(((((....)))))..))).))))...))))).)))) ( -35.70) >DroSec_CAF1 7335 104 - 1 UUCACCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGGGAGAAGGGUGAUUGGCCUAGUCAGGGCCAAUUUGUAAUUAAGCAGCACACUUUGGAGCUGCGGCAG .((((((.(((..((((((..........)))))).)))....))))))(((((((.....)))))))...........(((((.(......).)))))..... ( -42.20) >DroSim_CAF1 13161 104 - 1 UCCACCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCAAUUUGUAAUUAAGCAGCACACUUUGGAGCUGCGGCAG ..(((((.((((.((((((..........))))))..))))..)))))((((((((.....))))))))..........(((((.(......).)))))..... ( -41.80) >DroEre_CAF1 11548 94 - 1 --CUCCUGCUCGAGGCCAGGUUCAAGUUACUGGCCAGUGAGAAGGGUUAUUGGCCUACUCAGGGCCAAUUUGUAAUUAAGUACCACACUUUGGUAC-------- --.((((.((((.((((((..........))))))..)))).))))..((((((((.....))))))))..........((((((.....))))))-------- ( -36.40) >DroYak_CAF1 10828 102 - 1 --CUCCUUCUCGAGGCCAGGUUCGAGAUACUGGUCAGUGAGAAGGGUUAUUGGCCUAGUCAGGGCCAAUUUGUAAUUAAGUACCAUACUUUGAAACUGCGGCAG --.(((((((((.((((((..........))))))..)))))))))..((((((((.....)))))))).(((.....(((..((.....))..)))...))). ( -34.90) >consensus __CUCCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCAAUUUGUAAUUAAGUAGCACACUUUGGAGCUGCGGCAG ....(((.((((.((((((..........))))))..)))).)))..(((((((((.....))))))))).........(((((.(......).)))))..... (-30.40 = -31.96 + 1.56)

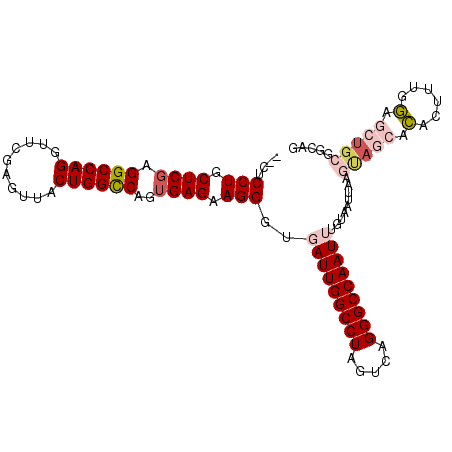
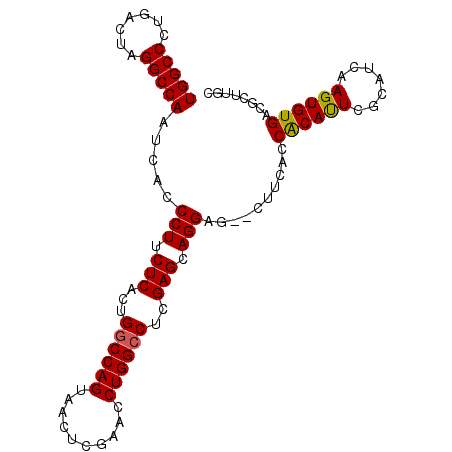
| Location | 1,255,655 – 1,255,753 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.74 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1255655 98 + 20766785 UGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGAG--CUUCACCGCAUUCGCAUCAAGUGUGACGCUUGU (((((.......))))).....(((.(((...((((((..........))))))..))).)))((--(.((((.((...........)))))).)))... ( -30.90) >DroSec_CAF1 7372 100 + 1 UGGCCCUGACUAGGCCAAUCACCCUUCUCCCUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGUGAAGCUCACCACACUCGCAUCAAGUGUGACGCUUGC (((((.......))))).(((((...(((...((((((..........))))))..)))..)))))(((....((((((.......))))))..)))... ( -34.80) >DroSim_CAF1 13198 100 + 1 UGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGUGGAGCUCACCACAUUCGCAUCAAGUGUGACGCUUGC (((((.......)))))..((((...(((...((((((..........))))))..)))..))))((((((((.((...........)))))).)))).. ( -33.60) >DroEre_CAF1 11577 93 + 1 UGGCCCUGAGUAGGCCAAUAACCCUUCUCACUGGCCAGUAACUUGAACCUGGCCUCGAGCAGGAG--CUGCACCACAUUCGCAUCAAGUGUGACG----- (((((.((((.(((........))).))))..)))))((.((((((.((((........))))..--.(((.........)))))))))...)).----- ( -29.00) >DroYak_CAF1 10865 98 + 1 UGGCCCUGACUAGGCCAAUAACCCUUCUCACUGACCAGUAUCUCGAACCUGGCCUCGAGAAGGAG--UUGCACCACAUUCGGAUCAAGUGUGACGCUUGC (((((.......)))))..((((((((((...(.((((..........)))).)..))))))).)--))((..((((((.......))))))..)).... ( -29.90) >consensus UGGCCCUGACUAGGCCAAUCACCCUUCUCACUGGCCAGUAACUCGAACCUGGCCUCGAGCAGGAG__CUUCACCACAUUCGCAUCAAGUGUGACGCUUGC (((((.......))))).....(((.(((...((((((..........))))))..))).)))..........((((((.......))))))........ (-27.86 = -27.74 + -0.12)

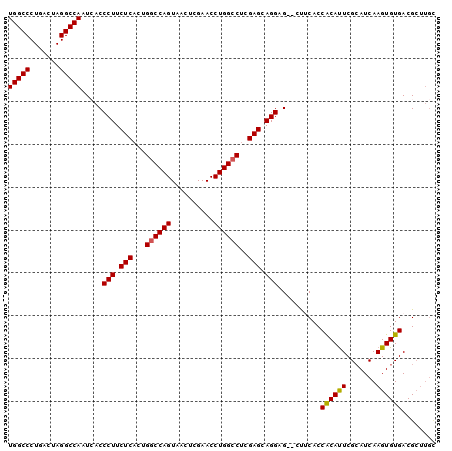
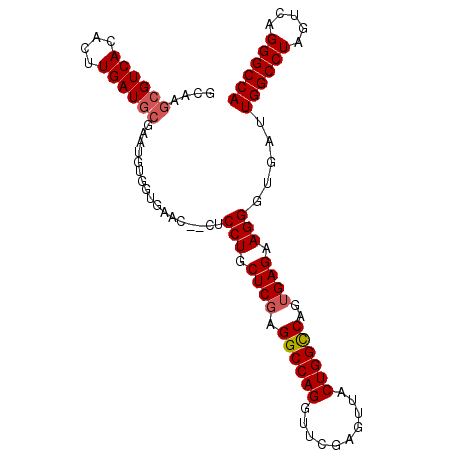
| Location | 1,255,655 – 1,255,753 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -39.24 |
| Consensus MFE | -31.46 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1255655 98 - 20766785 ACAAGCGUCACACUUGAUGCGAAUGCGGUGAAG--CUCCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCA ....((((((....))))))....((......)--)((((.((((.((((((..........))))))..)))).))))....((((((.....)))))) ( -37.00) >DroSec_CAF1 7372 100 - 1 GCAAGCGUCACACUUGAUGCGAGUGUGGUGAGCUUCACCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGGGAGAAGGGUGAUUGGCCUAGUCAGGGCCA ((...(..((((((((...))))))))..).)).((((((.(((..((((((..........)))))).)))....)))))).((((((.....)))))) ( -45.20) >DroSim_CAF1 13198 100 - 1 GCAAGCGUCACACUUGAUGCGAAUGUGGUGAGCUCCACCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCA (((.((((((....))))))...))).........(((((.((((.((((((..........))))))..))))..)))))..((((((.....)))))) ( -38.80) >DroEre_CAF1 11577 93 - 1 -----CGUCACACUUGAUGCGAAUGUGGUGCAG--CUCCUGCUCGAGGCCAGGUUCAAGUUACUGGCCAGUGAGAAGGGUUAUUGGCCUACUCAGGGCCA -----(..((((.(((...))).))))..).((--(.(((.((((.((((((..........))))))..)))).))))))..((((((.....)))))) ( -34.30) >DroYak_CAF1 10865 98 - 1 GCAAGCGUCACACUUGAUCCGAAUGUGGUGCAA--CUCCUUCUCGAGGCCAGGUUCGAGAUACUGGUCAGUGAGAAGGGUUAUUGGCCUAGUCAGGGCCA ....((..((((.(((...))).))))..))((--(.((((((((.((((((..........))))))..)))))))))))..((((((.....)))))) ( -40.90) >consensus GCAAGCGUCACACUUGAUGCGAAUGUGGUGAAC__CUCCUGCUCGAGGCCAGGUUCGAGUUACUGGCCAGUGAGAAGGGUGAUUGGCCUAGUCAGGGCCA ....((((((....)))))).................(((.((((.((((((..........))))))..)))).))).....((((((.....)))))) (-31.46 = -31.90 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:20 2006