| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
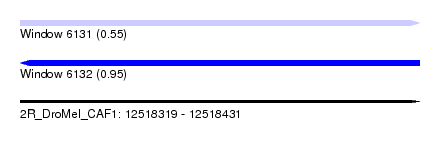
| Location | 12,518,319 – 12,518,431 |
| Length | 112 |
| Max. P | 0.952771 |

| Location | 12,518,319 – 12,518,431 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.57 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
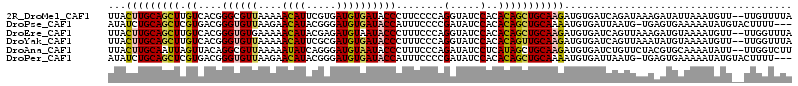
>2R_DroMel_CAF1 12518319 112 + 20766785 UUACUUGCAGCUUGUCACGGGCGUUAAAAACAUUCGUGAUGUGAUACCCUUCCCCAGGUAUCCACACAGCUGCAAGAUGUGAUCAGAUAAAGAUAUUAAAUGUU--UUGUUUUA ...(((((((((.((((((((.((.....)).))))))))((((((((........))))).)))..))))))))).............(((((((...)))))--))...... ( -32.30) >DroPse_CAF1 1742 110 + 1 AUAUCUGCAGCUCGUGACGGGUGUUAAGAACAUACGGGAUGUGAUACCAUUUCCCCGAUAUCCACACAGCUGCAAAAUGUGAUUAAUG-UGAGUGAAAAAUAUGUACUUUU--- .....(((((((.(((..(((((((..(......)((((.(((....))).)))).))))))).))))))))))....(((....(((-(.........)))).)))....--- ( -27.60) >DroEre_CAF1 1003 112 + 1 UUACUUGCAGCUUGUCACGGGUGUGAAAAACAUACGAGAUGUAAUACCCUUUCCCAGGUAUCCACACAGCUGCAAGAUGUGAUCAGUUAAAGAUGUAAAAUGUU--UUGGUUUA ...(((((((((.(((.((..(((.....)))..)).)))((.(((((........)))))..))..)))))))))....(((((...(((.((.....)).))--)))))).. ( -26.90) >DroYak_CAF1 1277 112 + 1 UUACUUGCAGCUUGUCACGGGUGUUAAAAACAUUCGCGAUGUGAUACCCUUUCCCAGGUAUCCACACAGUUGCAAGAUGUGAUCAGUUAAAUAUGUAAAAUGUU--UUGGUUUA ...(((((((((.(((.(((((((.....))))))).)))((((((((........))))).)))..)))))))))....(((((...((((((.....)))))--)))))).. ( -33.00) >DroAna_CAF1 1419 112 + 1 UUACUUGCAAUUAGUUACAGGCGUUAAAAAUAUCAGGGAUGUAAUACCCUUUCCCAGAUAUCCUCAUAGCUGCAAGAUGUGAUCUGUUCUACGUGCAAAAUAUU--UUGGUCUU ....(((((..(((..(((((........(((((.((((............)))).)))))..(((((.(.....).)))))))))).)))..)))))......--........ ( -21.90) >DroPer_CAF1 1760 110 + 1 AUAUCUGCAGCUCGUGACGGGUGUUAAGAACAUACGGGAUGUGAUACCAUUUCCCCGAUAUCCACACAGCUGCAAAAUGUGAUUAAUG-UGAGUGAAAAAUAUGUACUUUU--- .....(((((((.(((..(((((((..(......)((((.(((....))).)))).))))))).))))))))))....(((....(((-(.........)))).)))....--- ( -27.60) >consensus UUACUUGCAGCUUGUCACGGGUGUUAAAAACAUACGGGAUGUGAUACCCUUUCCCAGAUAUCCACACAGCUGCAAGAUGUGAUCAGUUAAAAAUGUAAAAUAUU__UUGGUUUA ...(((((((((.((....((((((....((((.....))))))))))........(.....)..)))))))))))...................................... (-15.65 = -15.57 + -0.08)
| Location | 12,518,319 – 12,518,431 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -21.85 |
| Energy contribution | -20.63 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
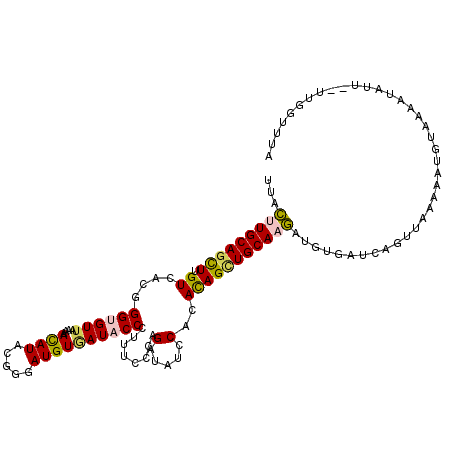
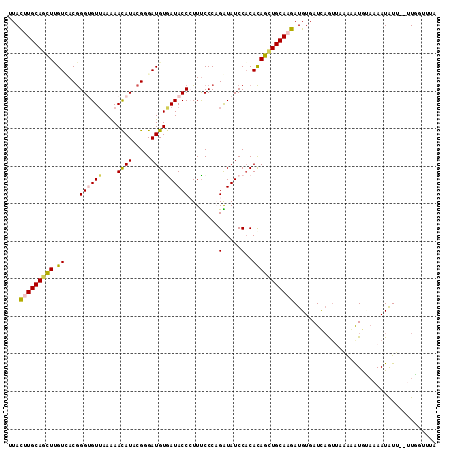
>2R_DroMel_CAF1 12518319 112 - 20766785 UAAAACAA--AACAUUUAAUAUCUUUAUCUGAUCACAUCUUGCAGCUGUGUGGAUACCUGGGGAAGGGUAUCACAUCACGAAUGUUUUUAACGCCCGUGACAAGCUGCAAGUAA ........--............................(((((((((.((((.((((((......))))))))))(((((..(((.....)))..)))))..)))))))))... ( -31.60) >DroPse_CAF1 1742 110 - 1 ---AAAAGUACAUAUUUUUCACUCA-CAUUAAUCACAUUUUGCAGCUGUGUGGAUAUCGGGGAAAUGGUAUCACAUCCCGUAUGUUCUUAACACCCGUCACGAGCUGCAGAUAU ---......................-............((((((((((((.((((((((((...(((......))))))).))))))...........))).)))))))))... ( -26.10) >DroEre_CAF1 1003 112 - 1 UAAACCAA--AACAUUUUACAUCUUUAACUGAUCACAUCUUGCAGCUGUGUGGAUACCUGGGAAAGGGUAUUACAUCUCGUAUGUUUUUCACACCCGUGACAAGCUGCAAGUAA ........--............................((((((((((((((.((((((......)))))))))))............((((....))))..)))))))))... ( -27.00) >DroYak_CAF1 1277 112 - 1 UAAACCAA--AACAUUUUACAUAUUUAACUGAUCACAUCUUGCAACUGUGUGGAUACCUGGGAAAGGGUAUCACAUCGCGAAUGUUUUUAACACCCGUGACAAGCUGCAAGUAA ........--............................((((((.((.((((.((((((......))))))))))(((((..(((.....)))..)))))..)).))))))... ( -25.30) >DroAna_CAF1 1419 112 - 1 AAGACCAA--AAUAUUUUGCACGUAGAACAGAUCACAUCUUGCAGCUAUGAGGAUAUCUGGGAAAGGGUAUUACAUCCCUGAUAUUUUUAACGCCUGUAACUAAUUGCAAGUAA ........--.....((((((..(((...((((...))))((((((..((((((((((.((((...(......).)))).))))))))))..).))))).)))..))))))... ( -25.90) >DroPer_CAF1 1760 110 - 1 ---AAAAGUACAUAUUUUUCACUCA-CAUUAAUCACAUUUUGCAGCUGUGUGGAUAUCGGGGAAAUGGUAUCACAUCCCGUAUGUUCUUAACACCCGUCACGAGCUGCAGAUAU ---......................-............((((((((((((.((((((((((...(((......))))))).))))))...........))).)))))))))... ( -26.10) >consensus UAAAACAA__AACAUUUUACACCUAUAACUGAUCACAUCUUGCAGCUGUGUGGAUACCUGGGAAAGGGUAUCACAUCCCGUAUGUUUUUAACACCCGUGACAAGCUGCAAGUAA ......................................((((((((((((((.((((((......)))))))))))..((..(((.....)))..)).....)))))))))... (-21.85 = -20.63 + -1.22)

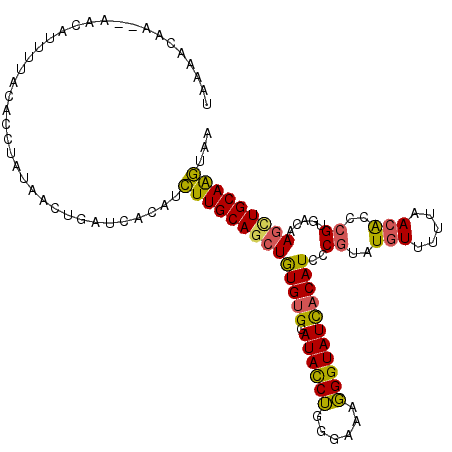
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:07 2006