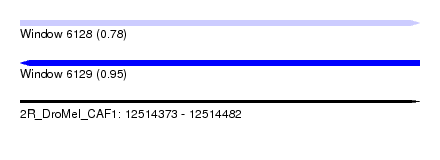
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,514,373 – 12,514,482 |
| Length | 109 |
| Max. P | 0.950126 |

| Location | 12,514,373 – 12,514,482 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.83 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -13.37 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12514373 109 + 20766785 UACACGAGGGCGACCACCACCAAGGUGGUAAUCAUCGCUCCCACAACCAGGUCAACGACAAAGUGACAGAUGCGGGACCACCACGGUGGUUUUGGCCUGGGUUGCAAGC .....(.((((((..(((((....))))).....)))))).).((((((((((((((.((..........)))).((((((....))))))))))))).)))))..... ( -39.30) >DroPse_CAF1 44011 109 + 1 AACACAAUCGAAACCACGAUCAAACUCAUGAUGAUUAUGGCCAGAAUGGUUUCCAGGAUCUCGCAGCCAAUCCGACGCGUCCACGACUGAGUCCCCCUCUUGUGUAGGU .((((((..(((((((...((....((((((...))))))...)).)))))))..(((.(((((..(......)..))(((...))).)))))).....)))))).... ( -21.70) >DroSec_CAF1 48773 109 + 1 AACACCAGGGCGACCACCACCAAGGUGGUAACCAUCGCUCCCACAACCAGGUCAACGACAAAGUGACAGAUGCGGGAGCACCACGGUGGUUUUGGCCUGGGUUGCAAAC (((.(((((.(((..((((((..(((((...)))))((((((.((.(...((((.(......))))).).)).)))))).....)))))).))).))))))))...... ( -47.90) >DroEre_CAF1 43636 109 + 1 UACACCAGGGCGACCACCACCACAGUGGUAACCGCCACUCCCACAAUCAGGUCAACGACAAAGUGACAGAUGCGGGACCACCACGGUGGCUUCGGCCUGGGUUGUAAGC (((((((((.(((...(((((...(((((....)))))((((.((.((..((((.(......))))).)))).)))).......)))))..))).)))))..))))... ( -43.40) >DroYak_CAF1 44491 109 + 1 UAAACCAGGGCGACCACCACCACGGUGGUAAUCACUACUCCCAAAAUCACGUCAACGACGAAGCAACAUAUGCGGGACCACCACGGUGGUUUCGGCCUGGGUUGCAAAC ....(((((.(((..((((((..(((((..........((((.......((((...))))..(((.....))))))))))))..)))))).))).)))))......... ( -43.80) >DroPer_CAF1 44026 109 + 1 AACACAAUCGAAACCACGAUCAAACUCAUGAUGAUUAUGGCCAGAAUGGUUUCCAGGAUCUCGCAGCCAAUCCGACGCGUCCACGACUGAGUCCCCCUCUUGUGCAGGU ..(((((..(((((((...((....((((((...))))))...)).)))))))..(((.(((((..(......)..))(((...))).)))))).....)))))..... ( -19.90) >consensus AACACCAGGGCGACCACCACCAAAGUGGUAAUCAUCACUCCCACAAUCAGGUCAACGACAAAGCAACAGAUGCGGGACCACCACGGUGGUUUCGGCCUGGGUUGCAAGC .........(((((((((((....)))))...........................................((((.((((....)))).)))).....)))))).... (-13.37 = -14.32 + 0.95)

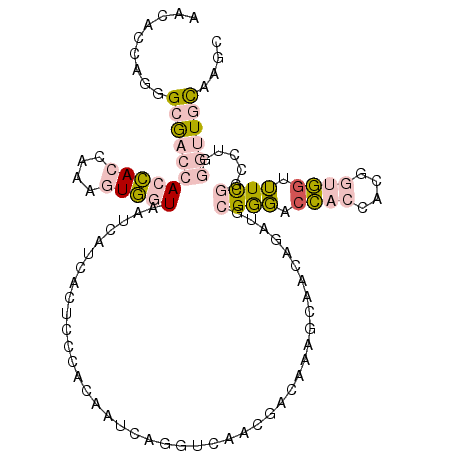
| Location | 12,514,373 – 12,514,482 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -24.45 |
| Energy contribution | -23.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12514373 109 - 20766785 GCUUGCAACCCAGGCCAAAACCACCGUGGUGGUCCCGCAUCUGUCACUUUGUCGUUGACCUGGUUGUGGGAGCGAUGAUUACCACCUUGGUGGUGGUCGCCCUCGUGUA .((..(((((.(((.(((...((..((((..(........)..))))..))...))).))))))))..)).((((((((((((((....))))))))))...))))... ( -43.00) >DroPse_CAF1 44011 109 - 1 ACCUACACAAGAGGGGGACUCAGUCGUGGACGCGUCGGAUUGGCUGCGAGAUCCUGGAAACCAUUCUGGCCAUAAUCAUCAUGAGUUUGAUCGUGGUUUCGAUUGUGUU ....((((((....(((((((.(((...)))(((((.....))).))))).)))).((((((((((..(((((.......))).))..))..))))))))..)))))). ( -31.20) >DroSec_CAF1 48773 109 - 1 GUUUGCAACCCAGGCCAAAACCACCGUGGUGCUCCCGCAUCUGUCACUUUGUCGUUGACCUGGUUGUGGGAGCGAUGGUUACCACCUUGGUGGUGGUCGCCCUGGUGUU ......(((((((((.(..(((((((.((((((((((((((.(((((......).))))..)).)))))))))...((...))))).)))))))..).).))))).))) ( -46.40) >DroEre_CAF1 43636 109 - 1 GCUUACAACCCAGGCCGAAGCCACCGUGGUGGUCCCGCAUCUGUCACUUUGUCGUUGACCUGAUUGUGGGAGUGGCGGUUACCACUGUGGUGGUGGUCGCCCUGGUGUA .......(((((((.(((.(((((((..(((((.((((..((.((((...((((......)))).)))).))..))))..)))))..)))))))..))).))))).)). ( -54.80) >DroYak_CAF1 44491 109 - 1 GUUUGCAACCCAGGCCGAAACCACCGUGGUGGUCCCGCAUAUGUUGCUUCGUCGUUGACGUGAUUUUGGGAGUAGUGAUUACCACCGUGGUGGUGGUCGCCCUGGUUUA .........(((((.(((.((((((((((((((((((((.....)))..((((...)))).......))))..........)))))))))))))..))).))))).... ( -44.90) >DroPer_CAF1 44026 109 - 1 ACCUGCACAAGAGGGGGACUCAGUCGUGGACGCGUCGGAUUGGCUGCGAGAUCCUGGAAACCAUUCUGGCCAUAAUCAUCAUGAGUUUGAUCGUGGUUUCGAUUGUGUU ....((((((....(((((((.(((...)))(((((.....))).))))).)))).((((((((((..(((((.......))).))..))..))))))))..)))))). ( -31.50) >consensus GCUUGCAACCCAGGCCGAAACCACCGUGGUGGUCCCGCAUCUGUCACUUUGUCGUUGACCUGAUUGUGGGAGUAAUGAUUACCACCUUGGUGGUGGUCGCCCUGGUGUA .........(((((.(((.(((((((.((((((((((((.(((...(.........)...))).)))))))..........))))).)))))))..))).))))).... (-24.45 = -23.90 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:04 2006