
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,495,275 – 12,495,422 |
| Length | 147 |
| Max. P | 0.987726 |

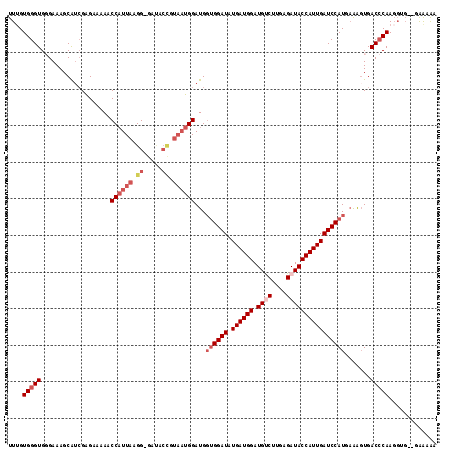
| Location | 12,495,275 – 12,495,385 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -20.92 |
| Energy contribution | -23.00 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12495275 110 + 20766785 UUUGUGGGUGGGAAAGCAUCGAGAAAAACCAUUAUGG-GAUACCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUGAGAUACCAUUGAUCCAUACAAGUGACCCAA------GAAAAA (((.(((((.(.................(((((((((-....)))))))))...((((((.((((((.((((....)))))))))))))))).....).))))).------)))... ( -36.40) >DroSec_CAF1 29864 108 + 1 UUUGUGGGUGGGAAAGCAUCGAGAAAAACCAUUAAGG-GA--------UGGAUGGUGGAUAUGAUGGAUGUCUUAAGAUACCAUUGAUCCAUUAAGGUGACCCAAGGUGAAGAAAAA ((..(...((((....((((........(((((....-))--------))).((((((((.((((((.((((....)))))))))))))))))).)))).))))..)..))...... ( -31.00) >DroSim_CAF1 25177 115 + 1 UUGGUGGGUGGGAAAGCAUCGAGAAAAACCAUUAAGG-GAUGCCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUAAGAUACCAUUGAUCCAUUAGGGUGACCCAAGGUGG-GAAAAA ((((((..(....)..))))))......(((((..((-(.((((.((((((((........((((((.((((....)))))))))))))))))).)))).)))..)))))-...... ( -35.70) >DroEre_CAF1 24985 114 + 1 UUUGUGGGUG-GAAAGCAUCGGGAA-AACCUUUAGGGCGCCACCGUAAUGGAUGCUGGAUAUGAUGGAUGCC-UGAGAUACCAUUGAUCCAUGAAAUUGACACAAGGUGUUCGGAAA ((((((((((-(...((...(((..-..))).....)).)))))...........(((((.((((((((...-....)).))))))))))).........))))))........... ( -27.50) >DroYak_CAF1 25492 114 + 1 UUUGUGGGUG-GAAAGCGGGGAGAA-AUCCAUUAAGG-GAUUCUGUAAUGGAUGGCGGAUUUGAUGGAUGCCUUGAGAUACCAUUGAUCCAUGAAAGUGACCCAAGGUGCAAGGAGA ....((((((-(....(((((....-((((((((((.-...(((((........))))))))))))))).))))).....)).......(((....))))))))............. ( -30.80) >consensus UUUGUGGGUGGGAAAGCAUCGAGAAAAACCAUUAAGG_GAUACCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUGAGAUACCAUUGAUCCAUGAAAGUGACCCAAGGUG__GAAAAA ....(((((...................((((((.((.....)).))))))...((((((.((((((.((((....)))))))))))))))).......)))))............. (-20.92 = -23.00 + 2.08)


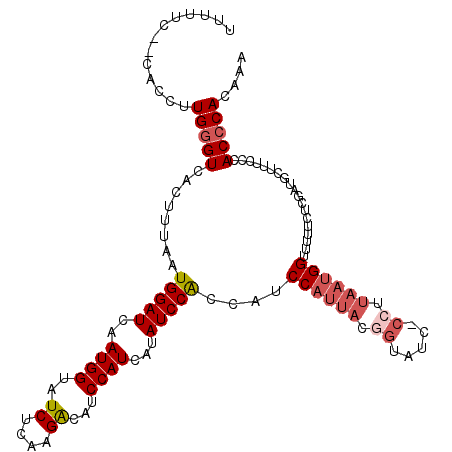
| Location | 12,495,275 – 12,495,385 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -15.58 |
| Energy contribution | -16.78 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12495275 110 - 20766785 UUUUUC------UUGGGUCACUUGUAUGGAUCAAUGGUAUCUCAAGACAUCCAUCAUAUCCACCAUCCAUUACGGUAUC-CCAUAAUGGUUUUUCUCGAUGCUUUCCCACCCACAAA ......------.(((((.....((((((((..((((..((....))...))))...)))).....((((((.((....-)).)))))).........))))......))))).... ( -23.10) >DroSec_CAF1 29864 108 - 1 UUUUUCUUCACCUUGGGUCACCUUAAUGGAUCAAUGGUAUCUUAAGACAUCCAUCAUAUCCACCAUCCA--------UC-CCUUAAUGGUUUUUCUCGAUGCUUUCCCACCCACAAA .............(((((.(((((((.((((..(((((....((.((......)).))...)))))..)--------))-).)))).))).......((.....))..))))).... ( -18.20) >DroSim_CAF1 25177 115 - 1 UUUUUC-CCACCUUGGGUCACCCUAAUGGAUCAAUGGUAUCUUAAGACAUCCAUCAUAUCCACCAUCCAUUACGGCAUC-CCUUAAUGGUUUUUCUCGAUGCUUUCCCACCCACCAA ......-......(((((........(((((..((((..((....))...))))...)))))....((((((.((....-)).))))))...................))))).... ( -21.30) >DroEre_CAF1 24985 114 - 1 UUUCCGAACACCUUGUGUCAAUUUCAUGGAUCAAUGGUAUCUCA-GGCAUCCAUCAUAUCCAGCAUCCAUUACGGUGGCGCCCUAAAGGUU-UUCCCGAUGCUUUC-CACCCACAAA .....(((.(((((((((((.....((((((...(((.......-((...)).......)))..)))))).....))))))....))))).-)))...........-.......... ( -22.54) >DroYak_CAF1 25492 114 - 1 UCUCCUUGCACCUUGGGUCACUUUCAUGGAUCAAUGGUAUCUCAAGGCAUCCAUCAAAUCCGCCAUCCAUUACAGAAUC-CCUUAAUGGAU-UUCUCCCCGCUUUC-CACCCACAAA .............(((((.........((((..((((..((....))...))))...))))...((((((((.((....-.))))))))))-..............-.))))).... ( -20.20) >consensus UUUUUC__CACCUUGGGUCACUUUAAUGGAUCAAUGGUAUCUCAAGACAUCCAUCAUAUCCACCAUCCAUUACGGUAUC_CCUUAAUGGUUUUUCUCGAUGCUUUCCCACCCACAAA .............(((((........(((((..((((..((....))...))))...)))))....((((((.((.....)).))))))...................))))).... (-15.58 = -16.78 + 1.20)

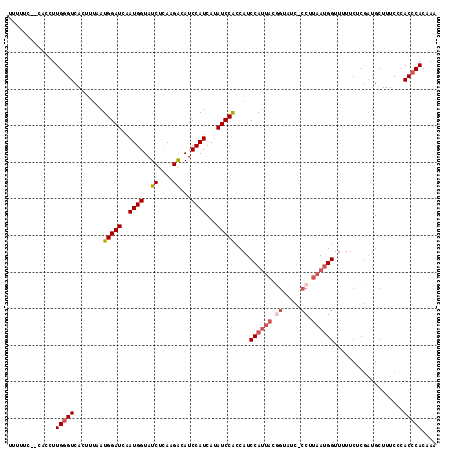
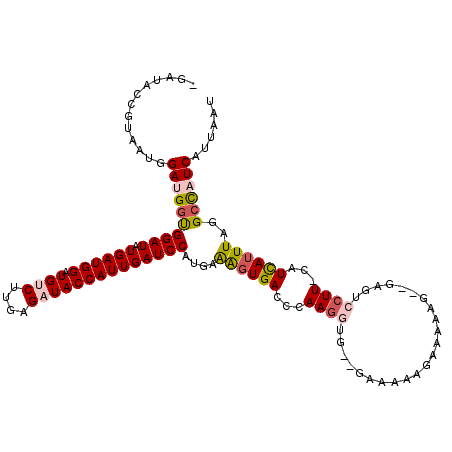
| Location | 12,495,312 – 12,495,422 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12495312 110 + 20766785 -GAUACCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUGAGAUACCAUUGAUCCAUACAAGUGACCCAA------GAAAAAGAAAAAG--GCGUCCUU-CAUCAUUUAGGCCAUCAUUAAU -(((.((......((((((((((.((((((.((((....))))))))))))))))..(((.(((((..------............)--).))))))-)))).....))..)))...... ( -27.14) >DroSec_CAF1 29901 110 + 1 -GA--------UGGAUGGUGGAUAUGAUGGAUGUCUUAAGAUACCAUUGAUCCAUUAAGGUGACCCAAGGUGAAGAAAAAGAAAAAAAAGGGUACUU-CAUUAUUUAGGCCAUCAUUAAU -((--------(((.((((((((.((((((.((((....))))))))))))))))))(((((.(((.......................))))))))-...........)))))...... ( -29.20) >DroSim_CAF1 25214 115 + 1 -GAUGCCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUAAGAUACCAUUGAUCCAUUAGGGUGACCCAAGGUGG-GAAAAAGAAAAAA--AGGUCCUU-CAUCAUUUAGGCCCUCAUUAAU -((.(((......(((((.((((..(((((.((((....)))))))))..((((((.((....))...)))))-)............--..)))).)-)))).....)))..))...... ( -32.60) >DroEre_CAF1 25020 112 + 1 CGCCACCGUAAUGGAUGCUGGAUAUGAUGGAUGCC-UGAGAUACCAUUGAUCCAUGAAAUUGACACAAGGUGUUCGGAAAGCAAGAG--GAGCCCUU-CAUCAUUUAUGCUUUCA----U ((.(((((((.....)))(((((.((((((((...-....)).)))))))))))..............))))..))(((((((.(((--((......-..)).))).))))))).----. ( -24.60) >DroYak_CAF1 25527 117 + 1 -GAUUCUGUAAUGGAUGGCGGAUUUGAUGGAUGCCUUGAGAUACCAUUGAUCCAUGAAAGUGACCCAAGGUGCAAGGAGACAAAGAG--GAGUCCUUCCAUCAUUUACGCUAUCAUUAAU -.......(((((.((((((....(((((((..(((((...((((.(((...(((....)))...)))))))))))).(((......--..)))..)))))))....))))))))))).. ( -36.40) >consensus _GAUACCGUAAUGGAUGGUGGAUAUGAUGGAUGUCUUGAGAUACCAUUGAUCCAUGAAAGUGACCCAAGGUG__GAAAAAGAAAAAG__GAGUCCUU_CAUCAUUUAGGCCAUCAUUAAU .............((((((((((.((((((.((((....))))))))))))))....((((((...((((.......................))))...))))))..))))))...... (-18.78 = -19.62 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:56 2006