
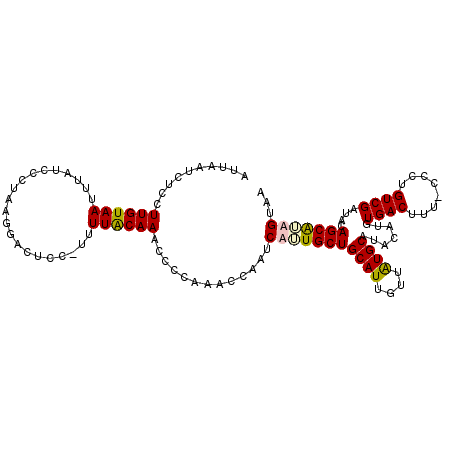
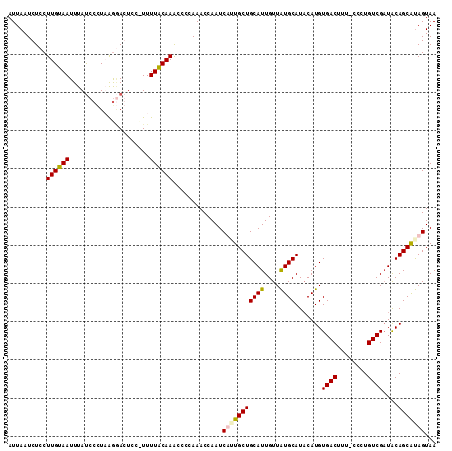
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,485,094 – 12,485,225 |
| Length | 131 |
| Max. P | 0.628048 |

| Location | 12,485,094 – 12,485,205 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12485094 111 + 20766785 AUUAAUCUCCUUGUAAUUUAUCCCUUAGGAGUCC-UUUUACAAACCCCAAACCAAUCAUUGCUGCAUUGUUAUGCAUACAUGUGACUUUUCCCUGUCGAUACAGCAUAGUAA ..........((((((....(((....)))....-..))))))................((((((((....)))).......((((........))))....))))...... ( -16.00) >DroSec_CAF1 19773 111 + 1 AUUAAUCUCCUUGUAAUUUAUCCCUUGGGAGUCC-UUUUACAAACCCCAAACCAAUCUUUGCUGCAUUGUUAUGCAUACAUGUGACUUUUCCCUGUCGAUGCAGCGUAGUAA ..........((((((....(((....)))....-..)))))).............((.(((((((((..((((....)))).(((........)))))))))))).))... ( -24.30) >DroEre_CAF1 14741 110 + 1 AUUAAUCUGCUUGUAAUUUACCCCUAAAAACUCCUUUUUGCAAACCCCAAACCAAUCAGUGCUGCAUUGUUGUGCAUACAUGUGACUU--CCCUGUCGAUACAGCACAGUAA .......(((((((((.....................))))))...............(((((((((....)))).......((((..--....))))....))))).))). ( -16.80) >DroYak_CAF1 15088 111 + 1 AUUAAUCUCCUUGUAAUUUAUCCUUAAGCACUCCCUUUUGCAAGCCCCAAACCAAUCUAUGCUGCAUUGUUAUGCAUUCAUGUGACUUU-UCCUGUCGCUACAGCAUAGUAA .........(((((((.....................)))))))............(((((((((((....))))......(((((...-....)))))...)))))))... ( -22.00) >consensus AUUAAUCUCCUUGUAAUUUAUCCCUAAGGACUCC_UUUUACAAACCCCAAACCAAUCAUUGCUGCAUUGUUAUGCAUACAUGUGACUUU_CCCUGUCGAUACAGCAUAGUAA ..........((((((.....................)))))).............(((((((((((....)))).......((((........))))....)))))))... (-13.98 = -14.22 + 0.25)

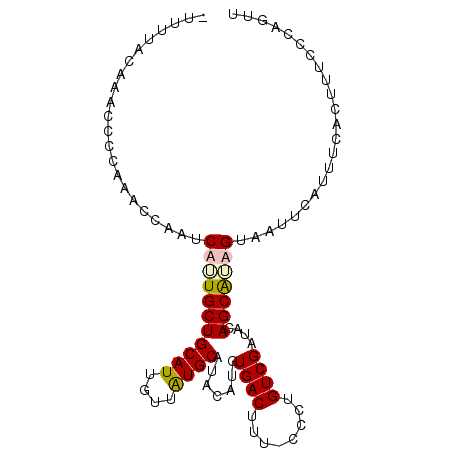
| Location | 12,485,128 – 12,485,225 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -15.31 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12485128 97 + 20766785 -UUUUACAAACCCCAAACCAAUCAUUGCUGCAUUGUUAUGCAUACAUGUGACUUUUCCCUGUCGAUACAGCAUAGUAAUUCAUUUCACUUUCCCAGUU -...........................((.((((((((((.......((((........)))).....)))))))))).))................ ( -10.50) >DroSec_CAF1 19807 97 + 1 -UUUUACAAACCCCAAACCAAUCUUUGCUGCAUUGUUAUGCAUACAUGUGACUUUUCCCUGUCGAUGCAGCGUAGUAAUUCAUUUCACUUUCCCAGUU -.....................((.(((((((((..((((....)))).(((........)))))))))))).))....................... ( -16.60) >DroEre_CAF1 14775 96 + 1 UUUUUGCAAACCCCAAACCAAUCAGUGCUGCAUUGUUGUGCAUACAUGUGACUU--CCCUGUCGAUACAGCACAGUAAUUCAUUUCACUUUCCCAUUU .......................((((.((.((((((((((.......((((..--....)))).....)))))))))).))...))))......... ( -15.30) >DroYak_CAF1 15122 97 + 1 CUUUUGCAAGCCCCAAACCAAUCUAUGCUGCAUUGUUAUGCAUUCAUGUGACUUU-UCCUGUCGCUACAGCAUAGUAAUUCAUUUCACUUUCAUACUU ....((.(((............(((((((((((....))))......(((((...-....)))))...)))))))............))).))..... ( -18.85) >consensus _UUUUACAAACCCCAAACCAAUCAUUGCUGCAUUGUUAUGCAUACAUGUGACUUU_CCCUGUCGAUACAGCAUAGUAAUUCAUUUCACUUUCCCAGUU ......................(((((((((((....)))).......((((........))))....)))))))....................... (-12.12 = -12.62 + 0.50)


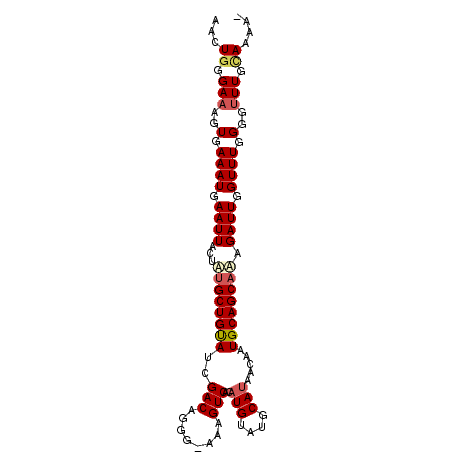
| Location | 12,485,128 – 12,485,225 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.85 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12485128 97 - 20766785 AACUGGGAAAGUGAAAUGAAUUACUAUGCUGUAUCGACAGGGAAAAGUCACAUGUAUGCAUAACAAUGCAGCAAUGAUUGGUUUGGGGUUUGUAAAA- ...((.(((..(.((((.(((((...((((((((.(((........)))..(((....)))....)))))))).))))).)))).)..))).))...- ( -19.80) >DroSec_CAF1 19807 97 - 1 AACUGGGAAAGUGAAAUGAAUUACUACGCUGCAUCGACAGGGAAAAGUCACAUGUAUGCAUAACAAUGCAGCAAAGAUUGGUUUGGGGUUUGUAAAA- ...((.(((..(.((((.((((.....(((((((.(((........)))..(((....)))....)))))))...)))).)))).)..))).))...- ( -18.80) >DroEre_CAF1 14775 96 - 1 AAAUGGGAAAGUGAAAUGAAUUACUGUGCUGUAUCGACAGGG--AAGUCACAUGUAUGCACAACAAUGCAGCACUGAUUGGUUUGGGGUUUGCAAAAA ...((.(((..(.((((.(((((..(((((((((.(((....--..)))...(((....)))...)))))))))))))).)))).)..))).)).... ( -24.20) >DroYak_CAF1 15122 97 - 1 AAGUAUGAAAGUGAAAUGAAUUACUAUGCUGUAGCGACAGGA-AAAGUCACAUGAAUGCAUAACAAUGCAGCAUAGAUUGGUUUGGGGCUUGCAAAAG .....((.((((.((((.(((..(((((((((((.(((....-...))).)(((....))).....))))))))))))).))))...)))).)).... ( -20.90) >consensus AACUGGGAAAGUGAAAUGAAUUACUAUGCUGUAUCGACAGGG_AAAGUCACAUGUAUGCAUAACAAUGCAGCAAAGAUUGGUUUGGGGUUUGCAAAA_ ...((.(((..(.((((.((((...((((((((..(((........)))..(((....))).....)))))))).)))).)))).)..))).)).... (-16.79 = -17.85 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:51 2006