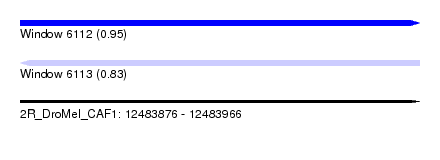
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,483,876 – 12,483,966 |
| Length | 90 |
| Max. P | 0.945128 |

| Location | 12,483,876 – 12,483,966 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945128 |
| Prediction | RNA |
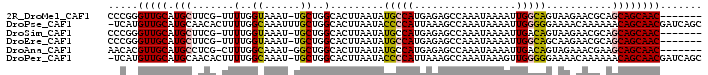
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12483876 90 + 20766785 -------GUUGCUGCUGCGUUCUUACUGCCAAUUUUAUUUGGCUCUCAUGGCAUAUUAAGUGCCAGCA-AUUUACCAAAA-CGAAGCAUGCAACCCGGG -------(((((((((.((((......(((((......))))).....((((((.....))))))...-.........))-)).)))).)))))..... ( -29.40) >DroPse_CAF1 14123 98 + 1 GCUGAUCGUUGCUGUUUUUUGUUUUCCCCCAAUUUUAUUUGGCUUUAAUGGGGUAUUAAGUGCCAGCAAAUUUGCCAAAAGUGUUGCAUGCAACAUGA- ((......((((((...((((...((((((((......)))).......))))...))))...))))))....)).....((((((....))))))..- ( -22.41) >DroSim_CAF1 13487 90 + 1 -------GUUGCUGCUGCGUUCUUACUGUCAAUUUUAUUUGGCUCUCAUGGCAUAUUAAGUGCCAGCA-AUUUACCAAAA-CGAAGCAUGCAACCCGGG -------(((((((((.((((......(((((......))))).....((((((.....))))))...-.........))-)).)))).)))))..... ( -26.70) >DroEre_CAF1 13546 90 + 1 -------GUUGCUGCUGCGUUCUUGCUGCCAAUUUUAUUUGGCUCUCAUGGCAUAUUAAGUGCCAGCA-AUUUACCAAAA-CGAAGCAUGCAACCCGGG -------(((((((((.((((.((((((((((......)))))......(((((.....)))))))))-)........))-)).)))).)))))..... ( -30.70) >DroAna_CAF1 14127 90 + 1 -------GUUGCUGCUUCGUUUCUACUGUCAAUUUUAUUUGGCUCUCAUGGCAUAUUAAGUGCCAGCC-AUUUGCCAAAG-CGAGGCAUGCAACGUGUU -------(((((((((((((.................((((((.....((((((.....))))))...-....)))))))-))))))).)))))..... ( -32.82) >DroPer_CAF1 14039 97 + 1 GCUGAUCGUUGCUGUUUUUUGUUUUCCCCCAACUUUAUUUGGCUUUAAUGGGGUAUUAAGUGCCAGCA-AUUUGCCAAAAGUGUUGCAUGCAACAUGA- ((.....(((((((...((((...((((((((......)))).......))))...))))...)))))-))..)).....((((((....))))))..- ( -21.61) >consensus _______GUUGCUGCUGCGUUCUUACUGCCAAUUUUAUUUGGCUCUCAUGGCAUAUUAAGUGCCAGCA_AUUUACCAAAA_CGAAGCAUGCAACCCGGG .......(((((((((.((........(((((......))))).....((((((.....))))))................)).)))).)))))..... (-19.34 = -19.70 + 0.36)

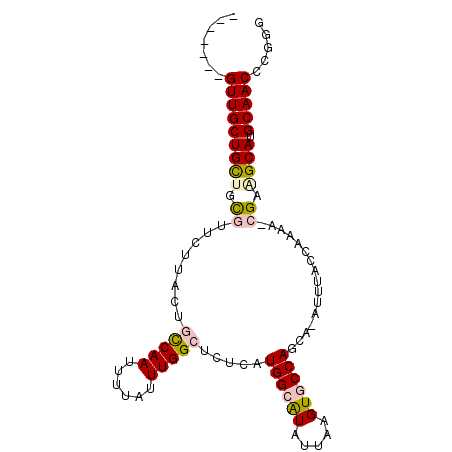
| Location | 12,483,876 – 12,483,966 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -14.85 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12483876 90 - 20766785 CCCGGGUUGCAUGCUUCG-UUUUGGUAAAU-UGCUGGCACUUAAUAUGCCAUGAGAGCCAAAUAAAAUUGGCAGUAAGAACGCAGCAGCAAC------- .....(((((.((((.((-((((.......-...(((((.......))))).....(((((......)))))....)))))).)))))))))------- ( -28.50) >DroPse_CAF1 14123 98 - 1 -UCAUGUUGCAUGCAACACUUUUGGCAAAUUUGCUGGCACUUAAUACCCCAUUAAAGCCAAAUAAAAUUGGGGGAAAACAAAAAACAGCAACGAUCAGC -...((((((...........(..(((....)))..).........(((((.................)))))..............))))))...... ( -19.63) >DroSim_CAF1 13487 90 - 1 CCCGGGUUGCAUGCUUCG-UUUUGGUAAAU-UGCUGGCACUUAAUAUGCCAUGAGAGCCAAAUAAAAUUGACAGUAAGAACGCAGCAGCAAC------- .....(((((.((((.((-((((.((.(((-(..((((.((((........)))).)))).....)))).))....)))))).)))))))))------- ( -25.90) >DroEre_CAF1 13546 90 - 1 CCCGGGUUGCAUGCUUCG-UUUUGGUAAAU-UGCUGGCACUUAAUAUGCCAUGAGAGCCAAAUAAAAUUGGCAGCAAGAACGCAGCAGCAAC------- .....(((((.((((.((-(((.......(-((((((((.......))))......(((((......))))))))))))))).)))))))))------- ( -30.21) >DroAna_CAF1 14127 90 - 1 AACACGUUGCAUGCCUCG-CUUUGGCAAAU-GGCUGGCACUUAAUAUGCCAUGAGAGCCAAAUAAAAUUGACAGUAGAAACGAAGCAGCAAC------- .....(((((.(((.((.-.((((((..((-(((.............)))))....))))))...........((....)))).))))))))------- ( -23.72) >DroPer_CAF1 14039 97 - 1 -UCAUGUUGCAUGCAACACUUUUGGCAAAU-UGCUGGCACUUAAUACCCCAUUAAAGCCAAAUAAAGUUGGGGGAAAACAAAAAACAGCAACGAUCAGC -...(((((....)))))...........(-(((((..........(((((.................)))))............))))))........ ( -19.18) >consensus CCCAGGUUGCAUGCUUCG_UUUUGGCAAAU_UGCUGGCACUUAAUAUGCCAUGAGAGCCAAAUAAAAUUGGCAGUAAAAACGAAGCAGCAAC_______ .....(((((.(((.......(..((......))..).........(((((.................)))))...........))))))))....... (-14.85 = -14.38 + -0.47)
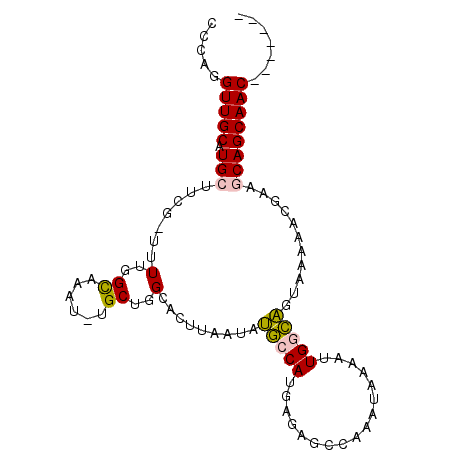
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:49 2006