
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,359,957 – 2,360,068 |
| Length | 111 |
| Max. P | 0.911999 |

| Location | 2,359,957 – 2,360,068 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -25.72 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
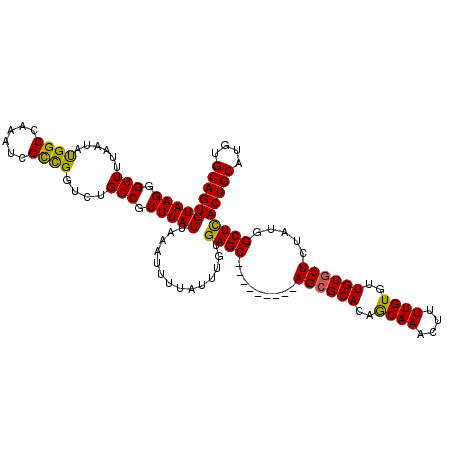
>2R_DroMel_CAF1 2359957 111 + 20766785 GUAAGGGUUUUAAUAUGGCCAAAUCGUCGGUCUGGCGUUUAUUAAAUUUUAUUUGUGAGC---------AGCGCACAACAAACUUUUGUGUUGUGCUGUAUGGCUCACUGCAUGUGCAGU (((...(((((((((..((((.((.....)).))))...)))))))........((((((---------.((((((((((........)))))))).))...)))))).))...)))... ( -33.30) >DroSec_CAF1 9417 111 + 1 GUAAGGGUUUUAAUAUGGCCAAAUCGCCGGUCUGGCGUUUAUUAAAUUUCAUUUGUGAGC---------AGCGCACAGCAAACUUUUGUGUUGUGCUCUAUGGCUCACUGCAUGUGCAGU ....(((.(((((((..((((.((.....)).))))...))))))).)))....((((((---------((.((((((((........)))))))).))...))))))(((....))).. ( -34.50) >DroSim_CAF1 9528 111 + 1 GUAAGGGUUUUAAUAUGGCCAAAUCGCCGGUCUGGCGUUUAUUAAAUUUCAUUUGUGAGC---------AGCGCACAGCAAACUUUUGUGUUGUGCUCUAUGGCUCACUGCAUUUGCAGU ....(((.(((((((..((((.((.....)).))))...))))))).)))....((((((---------((.((((((((........)))))))).))...))))))(((....))).. ( -34.50) >DroEre_CAF1 9551 111 + 1 GUAAGGGUUUUAAUACGGCGAAAUCGCUGCUCUGGCGUUUAUUAAAUUUUAUUUGCGAGC---------AGCGCACAGCAAACUUUUGUAUUGUGCUCUAUGGCUUACUGCAUGUGCAGU ((.(((((..(((((((((.....((((((((..(((..((........))..)))))))---------))))....)).......))))))).)))))...))..(((((....))))) ( -32.41) >DroYak_CAF1 8938 120 + 1 GUAAGGGUUUUAAUAUGGCGAACUCGCUGGUCUGGCGUUUAUUAAAUUUUAUUUGCGAGCACAGUUUGCAGGGCACAGCAAACUUUUGUAUUGUGCUGUAUGGCUUACUGCAUGUGCAGU (((((...(((((((.((((....)))).(.....)...))))))).....)))))..(((((...((((((((((((((.((....))....))))))...)))..))))))))))... ( -31.60) >consensus GUAAGGGUUUUAAUAUGGCCAAAUCGCCGGUCUGGCGUUUAUUAAAUUUUAUUUGUGAGC_________AGCGCACAGCAAACUUUUGUGUUGUGCUCUAUGGCUCACUGCAUGUGCAGU (((((.(((......((((......))))....))).)))))..............((((.........((((((..((((....))))..)))))).....))))(((((....))))) (-25.72 = -24.96 + -0.76)

| Location | 2,359,957 – 2,360,068 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -20.35 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
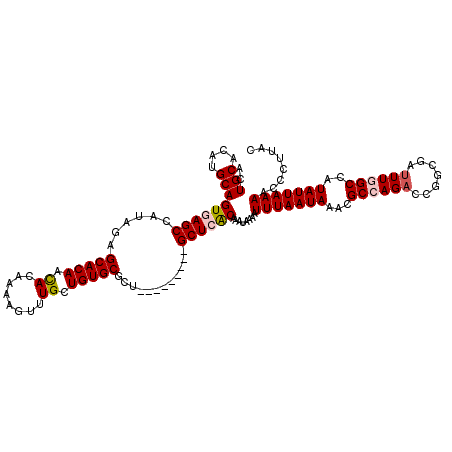
>2R_DroMel_CAF1 2359957 111 - 20766785 ACUGCACAUGCAGUGAGCCAUACAGCACAACACAAAAGUUUGUUGUGCGCU---------GCUCACAAAUAAAAUUUAAUAAACGCCAGACCGACGAUUUGGCCAUAUUAAAACCCUUAC ..(((....)))((((((......((((((((........))))))))...---------))))))........(((((((...((((((.......))))))..)))))))........ ( -31.90) >DroSec_CAF1 9417 111 - 1 ACUGCACAUGCAGUGAGCCAUAGAGCACAACACAAAAGUUUGCUGUGCGCU---------GCUCACAAAUGAAAUUUAAUAAACGCCAGACCGGCGAUUUGGCCAUAUUAAAACCCUUAC ..(((....)))((((((...((.(((((.((........)).))))).))---------))))))........(((((((...((((((.......))))))..)))))))........ ( -29.00) >DroSim_CAF1 9528 111 - 1 ACUGCAAAUGCAGUGAGCCAUAGAGCACAACACAAAAGUUUGCUGUGCGCU---------GCUCACAAAUGAAAUUUAAUAAACGCCAGACCGGCGAUUUGGCCAUAUUAAAACCCUUAC ..(((....)))((((((...((.(((((.((........)).))))).))---------))))))........(((((((...((((((.......))))))..)))))))........ ( -29.00) >DroEre_CAF1 9551 111 - 1 ACUGCACAUGCAGUAAGCCAUAGAGCACAAUACAAAAGUUUGCUGUGCGCU---------GCUCGCAAAUAAAAUUUAAUAAACGCCAGAGCAGCGAUUUCGCCGUAUUAAAACCCUUAC (((((....)))))..((......(((((..((....))....)))))(((---------((((((..................))..)))))))......))................. ( -25.67) >DroYak_CAF1 8938 120 - 1 ACUGCACAUGCAGUAAGCCAUACAGCACAAUACAAAAGUUUGCUGUGCCCUGCAAACUGUGCUCGCAAAUAAAAUUUAAUAAACGCCAGACCAGCGAGUUCGCCAUAUUAAAACCCUUAC ...((((((((((...((...((((((.(((......))))))))))).)))))...)))))............(((((((..(....)....(((....)))..)))))))........ ( -24.40) >consensus ACUGCACAUGCAGUGAGCCAUAGAGCACAACACAAAAGUUUGCUGUGCGCU_________GCUCACAAAUAAAAUUUAAUAAACGCCAGACCGGCGAUUUGGCCAUAUUAAAACCCUUAC ..(((....)))((((((......(((((.((........)).)))))............))))))........(((((((...((((((.......))))))..)))))))........ (-20.35 = -20.87 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:55 2006