| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,448,664 – 12,448,765 |
| Length | 101 |
| Max. P | 0.560423 |

| Location | 12,448,664 – 12,448,765 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12448664 101 - 20766785 U--------------GUG----AGUUAAGC-AUUUUCCAUGGGAAAAUUCAAUUGGAGGUUCCCAUGGCAACAGCAUCACACUUGGCAGCCCAGCUGAAUGUGACAUUUGAGUUGCCAAU (--------------(((----(.....((-..((.(((((((((..(((....)))..))))))))).))..)).))))).((((((((.(((.((.......)).))).)))))))). ( -36.40) >DroSec_CAF1 26901 101 - 1 A--------------GUA----AGUUAAGC-AUUUCCCAUGGGAAAAUUCAAUUGGAGGCUACCAUGGCAACAGCAUCACACUUGGCAGCCCAGCUGAAUGUGACAUUCGAGUUUCCAAU .--------------...----........-.(((((....))))).....((((((((((...(((((....)).(((((.(..((......))..).))))))))...)))))))))) ( -30.20) >DroSim_CAF1 27601 101 - 1 A--------------GUG----AGUUAAGC-AUUUUCCAGGGGAAAAUUCAAUUGGAGGCUACCAUGGCAACAGCAUCACACUUGGCAGCCCAGCUGAAUGUGACAUUCGAGUUUCCAAU .--------------...----......(.-(((((((...))))))).).((((((((((...(((((....)).(((((.(..((......))..).))))))))...)))))))))) ( -29.40) >DroEre_CAF1 22763 99 - 1 A--------------GUGAAAAUGCUCAGCUAUUUUCCAUGCGAAAAUUCAAUUGGAGUUUCCCAUGGCAGCAGCAUCACACCUGCCA-------UGAAUCUGCCAUUCGAGUUGCCAAU .--------------(.((((((((...)).)))))))....(..(((((...(((((.....((((((((...........))))))-------))...)).)))...)))))..)... ( -21.90) >DroYak_CAF1 45860 101 - 1 A--------------GCG----UGUUAAGC-AUUUUCCAUGGGAAAAUUCAAUUGAAGGUUCCCAUGGCUACAGCAUCACACUUGGCAGCCCAGUUGAAUGUGACAUUCGAGUUUCCAGU .--------------..(----(((...((-.....(((((((((..(((....)))..))))))))).....))...))))((((.(((....(((((((...)))))))))).)))). ( -31.50) >DroAna_CAF1 6224 111 - 1 UAAGAGGGUAGGUGGGGG----AGAUGGAA-AUU----GUAGGCAAAUUUAAUUCAAGGUUUCCAUGGACAGAGUAUCACACUUGAUGGCACAGAUGGAUGUGACAUUUGAGUUUCGAAU ...(((.((..((((..(----.((..(((-.((----(....))).)))...))....)..))))..))..........(((..(((.((((......)))).)).)..)))))).... ( -19.80) >consensus A______________GUG____AGUUAAGC_AUUUUCCAUGGGAAAAUUCAAUUGGAGGUUCCCAUGGCAACAGCAUCACACUUGGCAGCCCAGCUGAAUGUGACAUUCGAGUUUCCAAU ............................((......(((((((((..(((....)))..))))))))).....)).(((((.(((((......))))).)))))................ (-13.84 = -15.82 + 1.98)

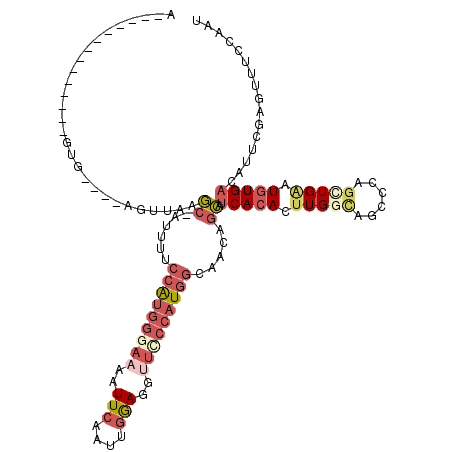

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:41 2006