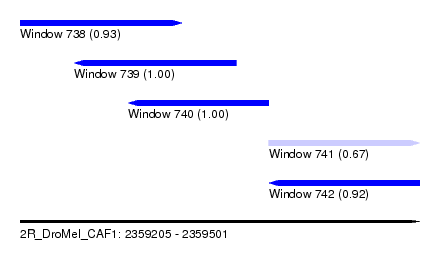
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,359,205 – 2,359,501 |
| Length | 296 |
| Max. P | 0.999822 |

| Location | 2,359,205 – 2,359,325 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -30.10 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2359205 120 + 20766785 CAAUCUGUUAACGCGUUUGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAUUUCGGGCGGCUCCCGGCCAUGUGUGCUCGCUCAAUCAAUUAUUCUGCCGAUUUCGGGUAUUUGCAUGU .................(((((((.........((((....))))((((((..((((..(((.....))).((.((....)).)).............))))..)))))))))))))... ( -32.30) >DroSec_CAF1 8683 120 + 1 CAAUCUGUUAACGCGUUUGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAUUUCGGGCGGCUCCCGGCCAUGUGUGCUCGCUCAAUCAAUUAUUCUGCCGAUUUCGGGUAUUUGCAUGU .................(((((((.........((((....))))((((((..((((..(((.....))).((.((....)).)).............))))..)))))))))))))... ( -32.30) >DroSim_CAF1 8807 120 + 1 CAAUCUGUUAACGCGUUUGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAUUUCGGGCGGCUCCCGGCCAUGUGUGCUCGCUCAAUCAAUUAUUCUGCCGAUUUCGGGUAUUUGCAUGU .................(((((((.........((((....))))((((((..((((..(((.....))).((.((....)).)).............))))..)))))))))))))... ( -32.30) >DroEre_CAF1 8836 120 + 1 CAAUCUGUUAACGCGUUGGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAUUUCGGGUGGCUCCCGGCCAUGUGUGCUCGCUCAAUCAAUUAUUCUGCCGAUUUCGGGUAUUUGCAUGU ((((.((....)).))))((((((.........((((....))))((((((..(((((((((.....)))))(((....)))................))))..)))))))))))).... ( -35.30) >DroYak_CAF1 8177 120 + 1 CAAUCUAUUAACGCGUUUGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAGUUCGUGUGGCUCCCGGCCAUGUGUGCCCGCUCAAUCAAUUAUUCUGCCGAUUUUGGGUAUUUGCGUGU ..........((((....((((((.........((((....))))(((((((.(((.(((((.....)))))(((....))).................))).))))))))))))))))) ( -27.00) >consensus CAAUCUGUUAACGCGUUUGCAAAUAAAAAUUAUCAAAUUAUUUUGGCCCGAUUUCGGGCGGCUCCCGGCCAUGUGUGCUCGCUCAAUCAAUUAUUCUGCCGAUUUCGGGUAUUUGCAUGU ..................((((((.........((((....))))((((((..(((.(((((.....))).((.((....)).))............)))))..)))))))))))).... (-30.10 = -29.70 + -0.40)

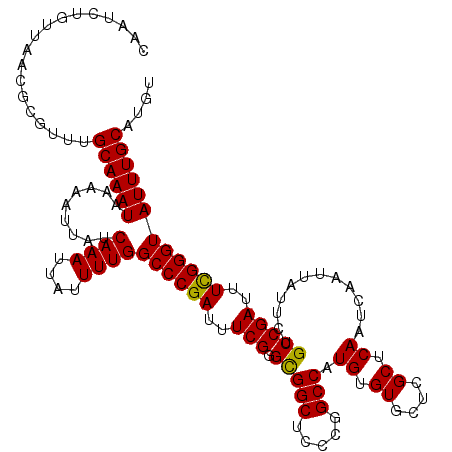
| Location | 2,359,245 – 2,359,365 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2359245 120 - 20766785 AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGCGAGCACACAUGGCCGGGAGCCGCCCGAAAUCGGGCCAAAA ..(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...))))).............(((.....)))(((((....)))))..... ( -35.40) >DroSec_CAF1 8723 120 - 1 AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGCGAGCACACAUGGCCGGGAGCCGCCCGAAAUCGGGCCAAAA ..(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...))))).............(((.....)))(((((....)))))..... ( -35.40) >DroSim_CAF1 8847 120 - 1 AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGCGAGCACACAUGGCCGGGAGCCGCCCGAAAUCGGGCCAAAA ..(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...))))).............(((.....)))(((((....)))))..... ( -35.40) >DroEre_CAF1 8876 120 - 1 AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGCGAGCACACAUGGCCGGGAGCCACCCGAAAUCGGGCCAAAA ..(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...)))))............((((((((.....)))(....).)))))... ( -34.00) >DroYak_CAF1 8217 120 - 1 AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACACGCAAAUACCCAAAAUCGGCAGAAUAAUUGAUUGAGCGGGCACACAUGGCCGGGAGCCACACGAACUCGGGCCAAAA ..(((((...((((((((((((.((((((((...)))))))).)))))))..((......))..)))))...)))))....(((.....((((.....))))..((....)).))).... ( -32.10) >consensus AGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGCGAGCACACAUGGCCGGGAGCCGCCCGAAAUCGGGCCAAAA ..................(((((.(((.((((((((((((....))))))..(((....)))........))))))..))).)))))..((((((((.....)))(....).)))))... (-31.76 = -32.16 + 0.40)

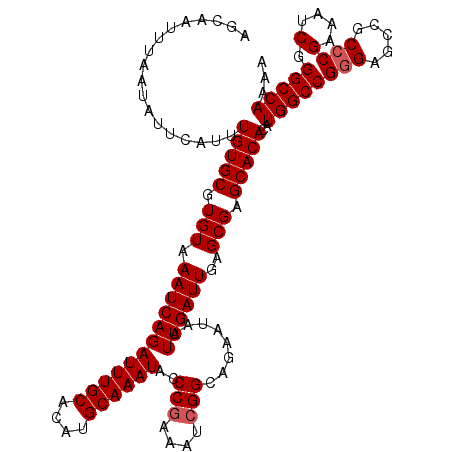
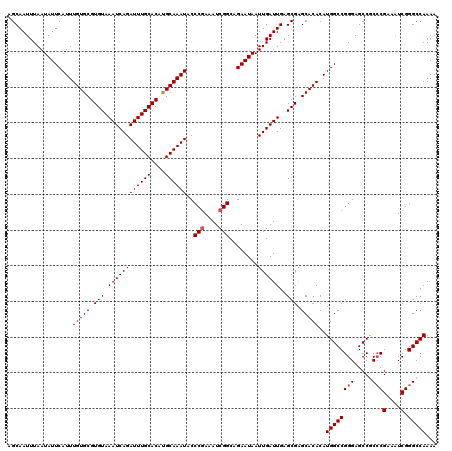
| Location | 2,359,285 – 2,359,389 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2359285 104 - 20766785 ----------------GUGUACUUGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGC ----------------.....((((((.((....))))))))(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...)))))... ( -26.80) >DroSec_CAF1 8763 104 - 1 ----------------GUGUACUUGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGC ----------------.....((((((.((....))))))))(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...)))))... ( -26.80) >DroSim_CAF1 8887 104 - 1 ----------------GUGUACUUGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGC ----------------.....((((((.((....))))))))(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...)))))... ( -26.80) >DroEre_CAF1 8916 104 - 1 ----------------GUGUACUUGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGC ----------------.....((((((.((....))))))))(((((...(((((((((((..((((((((...))))))))..))))))..(((....)))..)))))...)))))... ( -26.80) >DroYak_CAF1 8257 120 - 1 GUGUUUGGGUGUAUGGGUGUACUCGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACACGCAAAUACCCAAAAUCGGCAGAAUAAUUGAUUGAGC (.(((((((((........))))))))).)......(((((.((((....((((((((((((.((((((((...)))))))).)))))))..((......))..))))))))).))))). ( -31.50) >consensus ________________GUGUACUUGAACUCAAUUGAUUCAAGCAAUUUAAUAUUCAUUUGUGCGUGUAAAUCAGAUUUGCACAUGCAAAUACCCGAAAUCGGCAGAAUAAUUGAUUGAGC .....................((((((.((....))))))))(((((...((((((((((((.((((((((...)))))))).)))))))..(((....)))..)))))...)))))... (-25.10 = -25.34 + 0.24)

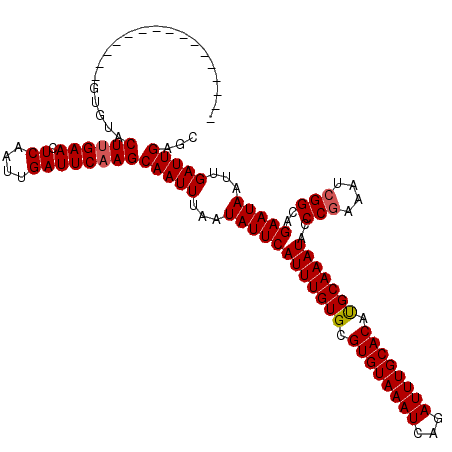
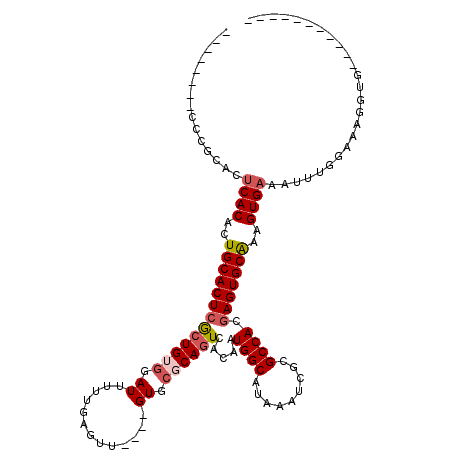
| Location | 2,359,389 – 2,359,501 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2359389 112 + 20766785 --------UCCGCACUCACCCUGCACUCGCUGUAGAUUUUUGAGUUCGCAGUGCUCAAUCACAAUGGCAUAAAUCGCGCCACGAGUGCAAAGUGAAAUUUGGAAAGGUGCCUUCCCGCAC --------...((..((((..((((((((.(((.(((...(((((.......))))))))))).((((.........))))))))))))..)))).....((.(((....))).)))).. ( -35.80) >DroSec_CAF1 8867 96 + 1 --------CCCGCACUCACACUGCACUCGCUGUGGAUUUUUGAGUU----GUGCGCAGUCACAAUGGCAUAAAUCGCGCCACGAGUGCAAAGUGAAAUUUGGAAAGG-G----------- --------(((.(..((((..((((((((..((((((((.((.(((----(((......))))))..)).))))).)))..))))))))..))))......)...))-)----------- ( -34.40) >DroSim_CAF1 8991 97 + 1 --------CCCGCACUCACACUGCACUCGCUGUGGAUUUUUGAGUU----GUGCGCAGUCACAAUGGCAUAAAUCGCGCCACAAGUGCAAAGUGAAAUUUGGAAAGGUG----------- --------.(((...((((..((((((...(((((.....(((.((----((((.((.......)))))))).)))..)))))))))))..))))....))).......----------- ( -28.60) >DroEre_CAF1 9020 94 + 1 --------CCAGCGCCCACACUGCACUCACUG-GGAUUUUUGAGUU----GUGCGCAGUCACAAUGGCA-AAACCGCGCCACGAGUGCGAAGUGAAAUUG-GAAAGCUG----------- --------.((((..(((((((((((((..((-((..(((((.(((----(((......))))))..))-)))...).))).))))))..))))....))-)...))))----------- ( -30.70) >DroYak_CAF1 8377 105 + 1 CCAAAUACCCAGCGCUCACACUGCACUCACUGUGGAUUUUUGAGUU----GUGCGCAGUCACAAUGGCAUAAAUCGCGCCACGAGUGCAAAGUGAAAUUGGGAAAGGUG----------- .......(((((...((((..(((((((...((((((((.((.(((----(((......))))))..)).))))).)))...)))))))..))))..))))).......----------- ( -36.20) >consensus ________CCCGCACUCACACUGCACUCGCUGUGGAUUUUUGAGUU____GUGCGCAGUCACAAUGGCAUAAAUCGCGCCACGAGUGCAAAGUGAAAUUUGGAAAGGUG___________ ...............((((..(((((((((((((.((.............)).)))))).....((((.........)))).)))))))..))))......................... (-21.64 = -22.44 + 0.80)

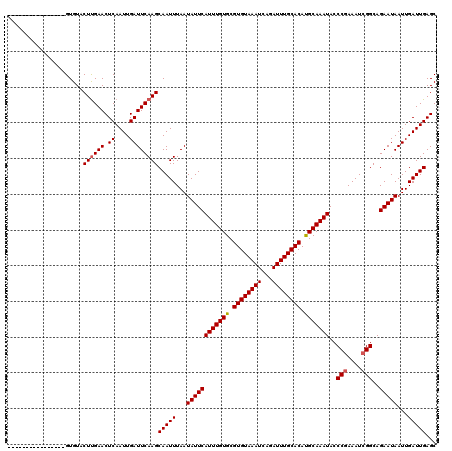
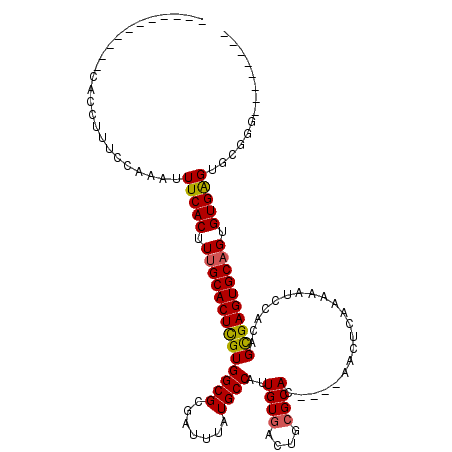
| Location | 2,359,389 – 2,359,501 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2359389 112 - 20766785 GUGCGGGAAGGCACCUUUCCAAAUUUCACUUUGCACUCGUGGCGCGAUUUAUGCCAUUGUGAUUGAGCACUGCGAACUCAAAAAUCUACAGCGAGUGCAGGGUGAGUGCGGA-------- (..(((((((....)))))).....(((((((((((((((((((.......))))..((((((((((.........))))...))).)))))))))))))))))))..)...-------- ( -45.50) >DroSec_CAF1 8867 96 - 1 -----------C-CCUUUCCAAAUUUCACUUUGCACUCGUGGCGCGAUUUAUGCCAUUGUGACUGCGCAC----AACUCAAAAAUCCACAGCGAGUGCAGUGUGAGUGCGGG-------- -----------.-.....((....(((((.((((((((((.(.(.(((((.((...(((((......)))----))..)).)))))).).)))))))))).)))))....))-------- ( -32.90) >DroSim_CAF1 8991 97 - 1 -----------CACCUUUCCAAAUUUCACUUUGCACUUGUGGCGCGAUUUAUGCCAUUGUGACUGCGCAC----AACUCAAAAAUCCACAGCGAGUGCAGUGUGAGUGCGGG-------- -----------.......((....(((((.((((((((((.(.(.(((((.((...(((((......)))----))..)).)))))).).)))))))))).)))))....))-------- ( -30.80) >DroEre_CAF1 9020 94 - 1 -----------CAGCUUUC-CAAUUUCACUUCGCACUCGUGGCGCGGUUU-UGCCAUUGUGACUGCGCAC----AACUCAAAAAUCC-CAGUGAGUGCAGUGUGGGCGCUGG-------- -----------((((..((-((....((((..(((((((((.(((((((.-.........))))))))))----.(((.........-.))))))))))))))))).)))).-------- ( -32.50) >DroYak_CAF1 8377 105 - 1 -----------CACCUUUCCCAAUUUCACUUUGCACUCGUGGCGCGAUUUAUGCCAUUGUGACUGCGCAC----AACUCAAAAAUCCACAGUGAGUGCAGUGUGAGCGCUGGGUAUUUGG -----------..((...((((..(((((.(((((((((..(.(.(((((.((...(((((......)))----))..)).)))))).)..))))))))).)))))...)))).....)) ( -34.80) >consensus ___________CACCUUUCCAAAUUUCACUUUGCACUCGUGGCGCGAUUUAUGCCAUUGUGACUGCGCAC____AACUCAAAAAUCCACAGCGAGUGCAGUGUGAGUGCGGG________ ........................(((((.((((((((((((((.......))))..((((....)))).....................)))))))))).))))).............. (-24.58 = -24.42 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:54 2006