
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,379,073 – 12,379,186 |
| Length | 113 |
| Max. P | 0.999937 |

| Location | 12,379,073 – 12,379,186 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.94 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.38 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12379073 113 + 20766785 UUGGAGGAUAAUGAUAAUCAUAUGCAAACACACAACCACCCAUCCGAUGAGGAGGGCCAUUACUAUGGC-------ACAUGAUUAUCAUUGCCUAAAAUCCUUUCCGAAAAAUUGAUCUC (((((((((((((((((((((.(((.............(((.(((.....)))))).(((....)))))-------).)))))))))))).......))))..)))))............ ( -32.11) >DroSec_CAF1 37988 114 + 1 UUGGAGGAAAAUGAUAAUCAUCUGAAAACACACAA---CCCAACCGAUGA---GGGCCAUUACUAUGGUUAUCAUAACAUGAUUAUCAUUGCCUAAAAUCCUUUCCGAAAAAUUGAUCUC ((((((((.((((((((((((.((........)).---........((((---..(((((....)))))..))))...))))))))))))........)))..)))))............ ( -25.10) >DroSim_CAF1 38481 107 + 1 UUGGAGGAAAAUGAUAAUCAUCUGCAAACAAACAA---CCCAUCCGAUGU---GGGCCAAUACUAUGAC-------ACAUGAUUAUCAUUGCCUAAAAUCCUCUCCGAAAAAUUGAUCUC ..((((((.((((((((((((.((((.........---(((((.....))---))).........)).)-------).))))))))))))........))))))................ ( -25.67) >DroEre_CAF1 37839 95 + 1 UUGGAGGUCAAUGAUUAUCAUCUGC-AACAUAUAA---CUUAUCCGAUAA---GGGCUACUACUAUUGC-------AGAUGAUUAUCAUUGGCUAAACCC-----------UUUGAUCUC ..((.((((((((((.(((((((((-((.......---....(((.....---))).........))))-------))))))).))))))))))...)).-----------......... ( -30.15) >DroYak_CAF1 38924 95 + 1 UCGGAGGAAAAUGACUAUCAUCUGA-AACGUAGAG---CCCAUCCAAUGA---CGGCCACUACUAUUGC-------AGAUGAUUAUCAUUGUCUAAACCC-----------AAUGAGUUU ..((.(((.(((((..((((((((.-...((((.(---((..((....))---.)))..)))).....)-------)))))))..))))).)))....))-----------......... ( -25.20) >consensus UUGGAGGAAAAUGAUAAUCAUCUGCAAACACACAA___CCCAUCCGAUGA___GGGCCACUACUAUGGC_______ACAUGAUUAUCAUUGCCUAAAAUCCU_UCCGAAAAAUUGAUCUC ....(((...(((((((((((......................((........))((((......)))).........)))))))))))..))).......................... (-10.62 = -11.94 + 1.32)

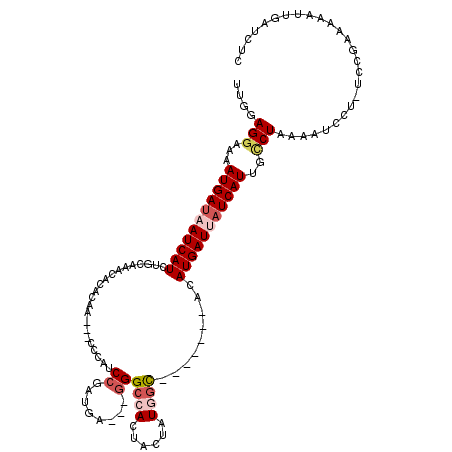
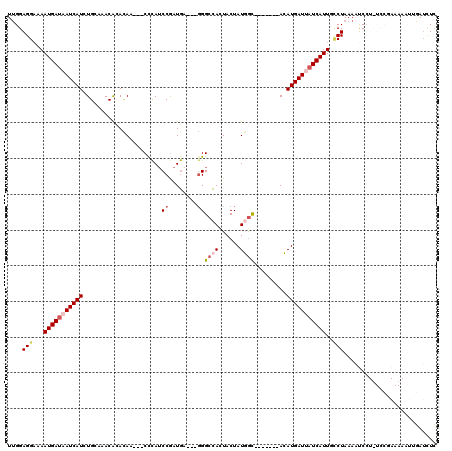
| Location | 12,379,073 – 12,379,186 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -14.36 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.42 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12379073 113 - 20766785 GAGAUCAAUUUUUCGGAAAGGAUUUUAGGCAAUGAUAAUCAUGU-------GCCAUAGUAAUGGCCCUCCUCAUCGGAUGGGUGGUUGUGUGUUUGCAUAUGAUUAUCAUUAUCCUCCAA ((((......))))(((..((((.......((((((((((((((-------((((((.((((.(((((((.....))).)))).))))))))...))))))))))))))))))))))).. ( -43.21) >DroSec_CAF1 37988 114 - 1 GAGAUCAAUUUUUCGGAAAGGAUUUUAGGCAAUGAUAAUCAUGUUAUGAUAACCAUAGUAAUGGCCC---UCAUCGGUUGGG---UUGUGUGUUUUCAGAUGAUUAUCAUUUUCCUCCAA ((((......))))(((..(((........((((((((((((....(((.((((((((...(((((.---.....)))))..---))))).))).))).)))))))))))).)))))).. ( -28.20) >DroSim_CAF1 38481 107 - 1 GAGAUCAAUUUUUCGGAGAGGAUUUUAGGCAAUGAUAAUCAUGU-------GUCAUAGUAUUGGCCC---ACAUCGGAUGGG---UUGUUUGUUUGCAGAUGAUUAUCAUUUUCCUCCAA ((((......))))((((.(((........((((((((((((.(-------((.((((...((((((---(.......))))---))).))))..))).))))))))))))))))))).. ( -31.80) >DroEre_CAF1 37839 95 - 1 GAGAUCAAA-----------GGGUUUAGCCAAUGAUAAUCAUCU-------GCAAUAGUAGUAGCCC---UUAUCGGAUAAG---UUAUAUGUU-GCAGAUGAUAAUCAUUGACCUCCAA (((.((...-----------.((.....))((((((.(((((((-------((((((...(((((((---.....))....)---)))).))))-))))))))).)))))))).)))... ( -32.10) >DroYak_CAF1 38924 95 - 1 AAACUCAUU-----------GGGUUUAGACAAUGAUAAUCAUCU-------GCAAUAGUAGUGGCCG---UCAUUGGAUGGG---CUCUACGUU-UCAGAUGAUAGUCAUUUUCCUCCGA (((((....-----------.))))).((.((((((.(((((((-------(.(((.((((.(.(((---((....))))).---).)))))))-.)))))))).)))))).))...... ( -34.70) >consensus GAGAUCAAUUUUUCGGA_AGGAUUUUAGGCAAUGAUAAUCAUGU_______GCCAUAGUAAUGGCCC___UCAUCGGAUGGG___UUGUAUGUUUGCAGAUGAUUAUCAUUUUCCUCCAA ((((((..............)))))).((.((((((((((((((.......(((((....)))))((........))....................)))))))))))))).))...... (-14.36 = -15.00 + 0.64)

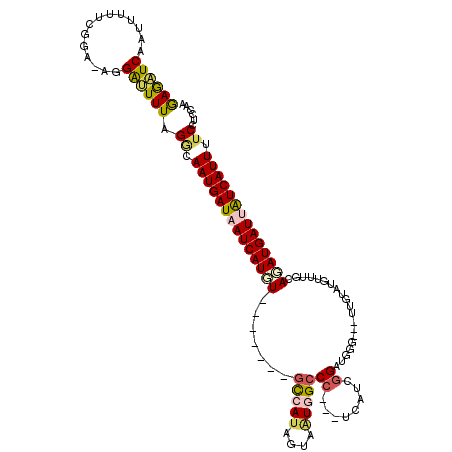
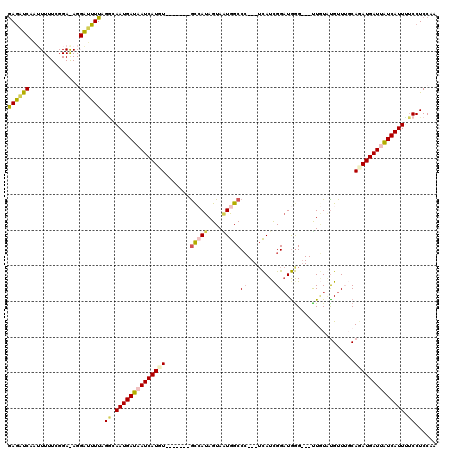
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:26 2006