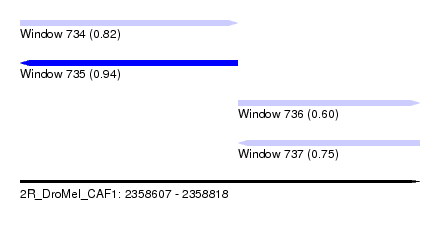
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,358,607 – 2,358,818 |
| Length | 211 |
| Max. P | 0.943254 |

| Location | 2,358,607 – 2,358,722 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.24 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -23.40 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2358607 115 + 20766785 AUAGGCAACAUUUGCCAUGAUAAUACGUAAUAUACAUAACAACCACAGUUAUGCGAGGCCGGGGGCGAUUCCGACCC-----UCAUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ...((((.....))))..................((((((.......)))))).((((..(((((((....)).)))-----))..((((((.....))))))...........)))).. ( -30.40) >DroSec_CAF1 8087 115 + 1 AUAGGAAACAUUUGCCAUGAUAAUACGUAAUAUACAUAACAACCACAGUUAUGCGAGGCCGGGGGCGAUACCGACCC-----CCAUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ..(((((..((((((((((......)))......((((((.......))))))...))).(((((((....)).)))-----))..((((((.....))))))...))))..)))))... ( -31.50) >DroSim_CAF1 8203 115 + 1 AUAGGCAACAUUUGCCAUGAUAAUACGUAAUAUACAUAACAACCACAGUUAUGCGAGGCCGGGGGCGAUACCGACCC-----CCAUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ...((((.....))))..................((((((.......)))))).((((..(((((((....)).)))-----))..((((((.....))))))...........)))).. ( -33.40) >DroEre_CAF1 8307 114 + 1 AUGGGCAACAUUUGCCAUGAUAAUACGUAAUACACAUAACAACCACAGUUAUGCCAGGCCGGAGGCGACUCCGACCCC------AUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ...((((.....((((((..(((((.(((.....((((((.......))))))...((.(((((....))))).))..------.))).)))))))))))))))................ ( -30.90) >DroYak_CAF1 7676 120 + 1 AAAGGCAACAUUUGCCAUGAUAAUACGUAAUAUACAUAACAAGCACAGUUAUGCGAGGCCGGAGGCAGCUCAAACCCCAUGCCCAUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ...((((.....((((((..(((((.(((.....((((((.......)))))).(.(((.((.((.........))))..)))).))).)))))))))))))))................ ( -25.50) >consensus AUAGGCAACAUUUGCCAUGAUAAUACGUAAUAUACAUAACAACCACAGUUAUGCGAGGCCGGGGGCGAUUCCGACCC_____CCAUGCAUAUUGAUGGUAUGCCGAAAAUAGUUCCUCAA ...((((.....((((((..(((((.(((.....((((((.......))))))...((.(((((....))))).)).........))).)))))))))))))))................ (-23.40 = -24.08 + 0.68)

| Location | 2,358,607 – 2,358,722 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.24 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2358607 115 - 20766785 UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAUGA-----GGGUCGGAAUCGCCCCCGGCCUCGCAUAACUGUGGUUGUUAUGUAUAUUACGUAUUAUCAUGGCAAAUGUUGCCUAU ..(((((....)))))((((..((((...(((((....-----(((((((........))))))).)))))...))))((((((((.(((........)))))))))))....))))... ( -33.80) >DroSec_CAF1 8087 115 - 1 UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAUGG-----GGGUCGGUAUCGCCCCCGGCCUCGCAUAACUGUGGUUGUUAUGUAUAUUACGUAUUAUCAUGGCAAAUGUUUCCUAU ...(((((.(((((.......(((((...(((((...(-----(((((((........)))))))))))))...))))).((((((.(((........)))))))))))))).))))).. ( -34.30) >DroSim_CAF1 8203 115 - 1 UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAUGG-----GGGUCGGUAUCGCCCCCGGCCUCGCAUAACUGUGGUUGUUAUGUAUAUUACGUAUUAUCAUGGCAAAUGUUGCCUAU ..(((((....)))))((((..((((...(((((...(-----(((((((........)))))))))))))...))))((((((((.(((........)))))))))))....))))... ( -35.60) >DroEre_CAF1 8307 114 - 1 UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAU------GGGGUCGGAGUCGCCUCCGGCCUGGCAUAACUGUGGUUGUUAUGUGUAUUACGUAUUAUCAUGGCAAAUGUUGCCCAU ..(((((....)))))((((..((((...(((((..------.(((((((((....))))))))).)))))...))))((((((((.(((........)))))))))))....))))... ( -38.50) >DroYak_CAF1 7676 120 - 1 UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAUGGGCAUGGGGUUUGAGCUGCCUCCGGCCUCGCAUAACUGUGCUUGUUAUGUAUAUUACGUAUUAUCAUGGCAAAUGUUGCCUUU ..(((((....)))))((((..(((((.....).))))(((((((((..(((....)))..)))))((((....))))((((((((.(((........)))))))))))))))))))... ( -32.30) >consensus UUGAGGAACUAUUUUCGGCAUACCAUCAAUAUGCAUGG_____GGGUCGGAAUCGCCCCCGGCCUCGCAUAACUGUGGUUGUUAUGUAUAUUACGUAUUAUCAUGGCAAAUGUUGCCUAU ..(((((....)))))((((..((((...(((((.........(((((((........))))))).)))))...))))((((((((.(((........)))))))))))....))))... (-28.36 = -28.64 + 0.28)

| Location | 2,358,722 – 2,358,818 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2358722 96 + 20766785 AGAUAAUAAAUAGAUGCCAAACGCAGUGGCCAGCGCUAGCAGCUCUGCUUUCGGCCAACAUA---------GCGCACUAAACACAGCCAGCUGCU-------G-CUGCUGUUA ..........(((..((((.......))))..(((((((((....)))....(.....).))---------)))).)))...(((((.(((....-------)-))))))).. ( -26.70) >DroSec_CAF1 8202 99 + 1 AGAUAAUAAAUAGAUGCCAAACGCAGUGGCCAGCGCCAGCAGCUCUGCUUUCGGCCAACAGA---------GCGCACUAAACACAGCCGGCUGCUGC----UG-CUGCUGUUA ...................(((((((..((.((((((.((.((((((...........))))---------))(.......)...)).))).)))))----..-)))).))). ( -32.70) >DroSim_CAF1 8318 102 + 1 AGAUAAUAAAUAGAUGCCGAACGCAGUGGCCAGCGCCAGCAGCUCUGCUUUCGGCCAACAGA---------GCGCACUAAACACAGCCGGCUGCUGCUG-CUG-CUGCUGUUA ......................((((..((.((((.((((((((..(((((.(.....))))---------))((..........)).)))))))).))-)))-)..)))).. ( -34.80) >DroEre_CAF1 8421 113 + 1 AGAUAAUAAAUAGCUGCCAAACGCAGUGGCCAGCGCCAUCAGCUCUGGAUUUGGCCAGCGGAGAGCGCUGUGCGCACUAAACACAGCCGGCCGCUGUUAACUGUCCACUGUUA .((((.(.((((((.(((...(((..(((((((..(((.......)))..))))))))))....(.((((((.........)))))))))).)))))).).))))........ ( -40.40) >consensus AGAUAAUAAAUAGAUGCCAAACGCAGUGGCCAGCGCCAGCAGCUCUGCUUUCGGCCAACAGA_________GCGCACUAAACACAGCCGGCUGCUGC____UG_CUGCUGUUA ........(((((.........(((((((((.(((((((((....))))...)))................(.(.......).).)).)))))))))..........))))). (-22.71 = -22.84 + 0.13)

| Location | 2,358,722 – 2,358,818 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.00 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2358722 96 - 20766785 UAACAGCAG-C-------AGCAGCUGGCUGUGUUUAGUGCGC---------UAUGUUGGCCGAAAGCAGAGCUGCUAGCGCUGGCCACUGCGUUUGGCAUCUAUUUAUUAUCU ...((((((-(-------(((.((..((.((((.....))))---------...))..))(....)....))))))...))))((((.......))))............... ( -31.90) >DroSec_CAF1 8202 99 - 1 UAACAGCAG-CA----GCAGCAGCCGGCUGUGUUUAGUGCGC---------UCUGUUGGCCGAAAGCAGAGCUGCUGGCGCUGGCCACUGCGUUUGGCAUCUAUUUAUUAUCU ...((((..-((----(((((.((((((.((((.....))))---------...))))))(....)....)))))))..))))((((.......))))............... ( -38.00) >DroSim_CAF1 8318 102 - 1 UAACAGCAG-CAG-CAGCAGCAGCCGGCUGUGUUUAGUGCGC---------UCUGUUGGCCGAAAGCAGAGCUGCUGGCGCUGGCCACUGCGUUCGGCAUCUAUUUAUUAUCU ...((((((-.((-(.(((((.....)))))((.....))))---------)))))))((((((.((((.(((((....)).)))..)))).))))))............... ( -39.30) >DroEre_CAF1 8421 113 - 1 UAACAGUGGACAGUUAACAGCGGCCGGCUGUGUUUAGUGCGCACAGCGCUCUCCGCUGGCCAAAUCCAGAGCUGAUGGCGCUGGCCACUGCGUUUGGCAGCUAUUUAUUAUCU ....((((((.((((..((((((..((((((((.......))))))).)...))))))(((((((.(((......((((....))))))).))))))))))).)))))).... ( -42.30) >consensus UAACAGCAG_CA____GCAGCAGCCGGCUGUGUUUAGUGCGC_________UCUGUUGGCCGAAAGCAGAGCUGCUGGCGCUGGCCACUGCGUUUGGCAUCUAUUUAUUAUCU ...((((((..........((.((..((((....))))))))..........))))))((((((.((((.(((((....)).)))..)))).))))))............... (-28.38 = -28.00 + -0.38)

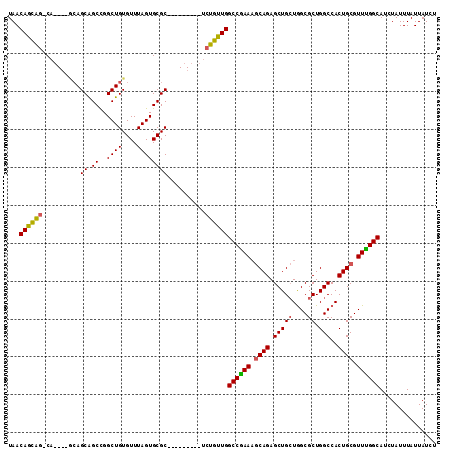
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:49 2006