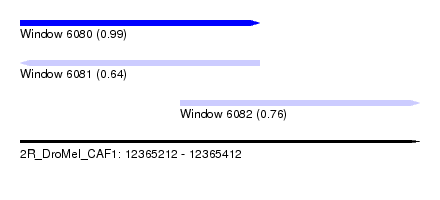
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,365,212 – 12,365,412 |
| Length | 200 |
| Max. P | 0.994214 |

| Location | 12,365,212 – 12,365,332 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12365212 120 + 20766785 GAGUUGGCAGGGAUGUAUCGAUAACACAAAUCAAAUUAUUUGGCAAAUUUAUCGAAAUCAAAAGCGGACAGAAUCUAUCGCCCAUCGAUAGAUACAUCGAUUGUAUAGCCACAGCCAGCU .(((((((...(((((((((((((..(((((......)))))......)))))))..................(((((((.....)))))))))))))...(((......)))))))))) ( -31.30) >DroSec_CAF1 24058 118 + 1 GAGUUGGCAGGGCUG--UCAAUAACAGAAAUCAGAUUAUUUGGCACAAGUAUCGAAACCAAAAGCGGACAAAAUCUAUCGCCCGUCGAUAGAUAAAUCGAUUGUAUGGCCACAGCCAGCU .(((((((.((.(((--(.....))))...))........((((......(((((..((......)).....((((((((.....))))))))...)))))......))))..))))))) ( -35.30) >DroSim_CAF1 24506 118 + 1 GAGUUGGCAGGGCUG--UCGAUAACAGAAAACAGAUUAUUUGGCACAAGUAUCGAAACCAAAAGCGGACAAAAUCUAUCGCCCAUCGAUAGAUAAAUCGAUUGUAUGGCCACAGCCAGCU .(((((((....(((--(.....)))).............((((......(((((..((......)).....((((((((.....))))))))...)))))......))))..))))))) ( -34.40) >DroEre_CAF1 24251 109 + 1 GAGUUGGCAGGGCAG--UCGAUAACAUGG---CGAUUGACUGG--CAAAUAUCGGCAUCAGAAGCUGACGAAACAUAUCGCCAAUCGA----UUCAUCGAUUGCAAAGCCACAGCCAGCU .(((((((..(.(((--(((((.......---..)))))))).--).......(((.((((...))))...........((.((((((----....))))))))...)))...))))))) ( -36.60) >DroYak_CAF1 24999 114 + 1 GAGUUGGCAGGGCAG--UCGAUAACAUAAAUCCGAUGAAUGGGCGCUAGUAUCGGUAUCAGAAGGUGAUAGUACAUAUCGCCAAUCGA----UACAUCGAUUGAAAAGCCACAGCCAGCU .(((((((..(((((--(((((..........(((((...(....)...)))))(((((.((.(((((((.....)))))))..))))----)))))))))).....)))...))))))) ( -38.50) >consensus GAGUUGGCAGGGCUG__UCGAUAACAGAAAUCAGAUUAUUUGGCACAAGUAUCGAAAUCAAAAGCGGACAAAAUCUAUCGCCCAUCGAUAGAUACAUCGAUUGUAUAGCCACAGCCAGCU .(((((((..(((....((((((........(((.....))).......))))))............................((((((......))))))......)))...))))))) (-21.28 = -21.52 + 0.24)

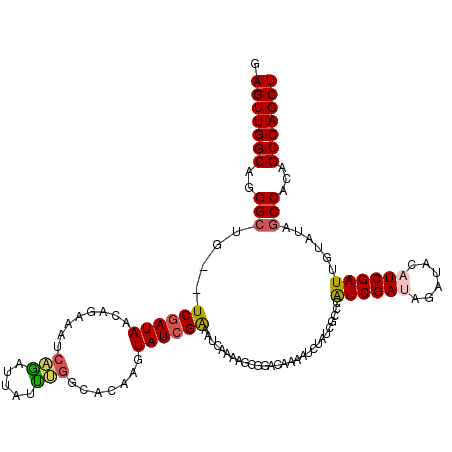
| Location | 12,365,212 – 12,365,332 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -19.51 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12365212 120 - 20766785 AGCUGGCUGUGGCUAUACAAUCGAUGUAUCUAUCGAUGGGCGAUAGAUUCUGUCCGCUUUUGAUUUCGAUAAAUUUGCCAAAUAAUUUGAUUUGUGUUAUCGAUACAUCCCUGCCAACUC ((.((((((((....))))...((((((((((((((.(((((((((...)))).)))))......)))))).....(((((((......))))).))....))))))))...)))).)). ( -30.70) >DroSec_CAF1 24058 118 - 1 AGCUGGCUGUGGCCAUACAAUCGAUUUAUCUAUCGACGGGCGAUAGAUUUUGUCCGCUUUUGGUUUCGAUACUUGUGCCAAAUAAUCUGAUUUCUGUUAUUGA--CAGCCCUGCCAACUC .((.(((((((((((....(((((...(((....(.(((((((......))))))))....))).)))))...)).))).((((((..(....).)))))).)--)))))..))...... ( -33.90) >DroSim_CAF1 24506 118 - 1 AGCUGGCUGUGGCCAUACAAUCGAUUUAUCUAUCGAUGGGCGAUAGAUUUUGUCCGCUUUUGGUUUCGAUACUUGUGCCAAAUAAUCUGUUUUCUGUUAUCGA--CAGCCCUGCCAACUC .((.((((((.(((.....((((((......)))))).)))((((((...........((((((..(((...))).))))))..........))))))....)--)))))..))...... ( -32.00) >DroEre_CAF1 24251 109 - 1 AGCUGGCUGUGGCUUUGCAAUCGAUGAA----UCGAUUGGCGAUAUGUUUCGUCAGCUUCUGAUGCCGAUAUUUG--CCAGUCAAUCG---CCAUGUUAUCGA--CUGCCCUGCCAACUC .........((((...(((.(((((((.----..(((((((((.(((((.((((((...))))))..))))))))--)))))).....---.....)))))))--.)))...)))).... ( -35.52) >DroYak_CAF1 24999 114 - 1 AGCUGGCUGUGGCUUUUCAAUCGAUGUA----UCGAUUGGCGAUAUGUACUAUCACCUUCUGAUACCGAUACUAGCGCCCAUUCAUCGGAUUUAUGUUAUCGA--CUGCCCUGCCAACUC ((.((((.(.(((...((((((((....----))))))))(((((.....(((((.....)))))(((((..............)))))........))))).--..)))).)))).)). ( -29.54) >consensus AGCUGGCUGUGGCUAUACAAUCGAUGUAUCUAUCGAUGGGCGAUAGAUUUUGUCCGCUUUUGAUUUCGAUACUUGUGCCAAAUAAUCUGAUUUAUGUUAUCGA__CAGCCCUGCCAACUC ((.((((.(.(((......((((((......))))))((((((......))))))...........(((((..........................))))).....)))).)))).)). (-19.51 = -20.11 + 0.60)


| Location | 12,365,292 – 12,365,412 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -27.32 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12365292 120 + 20766785 CCCAUCGAUAGAUACAUCGAUUGUAUAGCCACAGCCAGCUGGUCUCCCUUCGCCAUUCUUUAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUUAAGCC ...((((....(((((.....)))))........(((((((((((((((((((((.((...(((....)))..)).)))))))....))..))))))))).))).))))........... ( -31.10) >DroSec_CAF1 24136 120 + 1 CCCGUCGAUAGAUAAAUCGAUUGUAUGGCCACAGCCAGCUGGUCUCCCUUCGCCAUUCUUAAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUUAAGCC ..((((.((................((((....))))((((((((((((((((((.((..((((....)))).)).)))))))....))..))))))))))).))))............. ( -34.10) >DroSim_CAF1 24584 120 + 1 CCCAUCGAUAGAUAAAUCGAUUGUAUGGCCACAGCCAGCUGGUCUCCCUUCGCCAUUCUUUAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUUAAGCC ...(((....))).....((((((.((((....))))((((((((((((((((((.((...(((....)))..)).)))))))....))..)))))))))....)))))).......... ( -33.30) >DroEre_CAF1 24324 112 + 1 CCAAUCGA----UUCAUCGAUUGCAAAGCCACAGCCAGCUGGUCUCCCUUCGCC----UCUAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUCAAGCC .(((((((----....)))))))..........(((((((((((((((((((((----..(((........)))...))))))....))..))))))))).))).).............. ( -28.30) >DroYak_CAF1 25077 116 + 1 CCAAUCGA----UACAUCGAUUGAAAAGCCACAGCCAGCUGGUCUCCCUUCGCCAUUCUUUAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUCAAGCC .(((((((----....)))))))..........((((((((((((((((((((((.((...(((....)))..)).)))))))....))..))))))))).))).).............. ( -33.10) >consensus CCCAUCGAUAGAUACAUCGAUUGUAUAGCCACAGCCAGCUGGUCUCCCUUCGCCAUUCUUUAAUUUUUGUUUUGAUUGGUGAAUUUUGGCAGGGAUUAGCAUGGACGAUCCUCUUAAGCC ...((((((......))))))............((((((((((((((((((((((.((...(((....)))..)).)))))))....))..))))))))).))).).............. (-27.32 = -28.16 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:18 2006