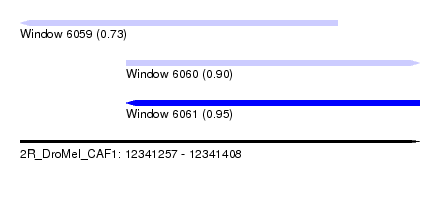
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,341,257 – 12,341,408 |
| Length | 151 |
| Max. P | 0.948702 |

| Location | 12,341,257 – 12,341,377 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -49.03 |
| Consensus MFE | -24.42 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
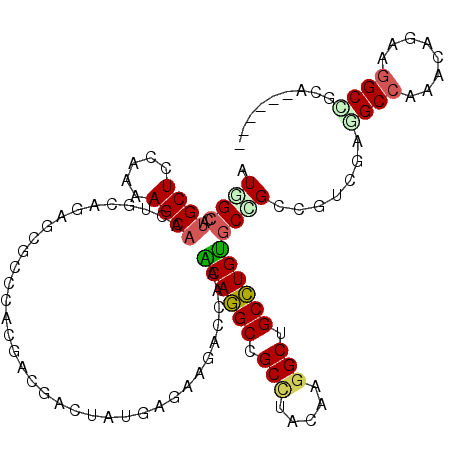
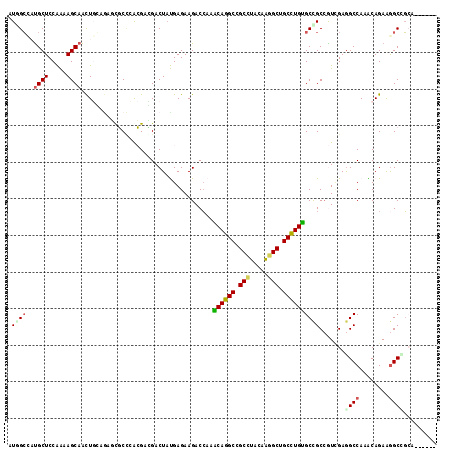
>2R_DroMel_CAF1 12341257 120 - 20766785 CAGGCAGCCUUGUAGGCGGCCUGCUUGGUCUUCUCAUAGUCGUCGUGGGCACUCACCAGUUGCUUUUGGAGCAUGGCCAUCCGGUUGCGAGCCGCUUCCAGGAGCUGUGUCUUGGCGGCC (((((.(((.....))).)))))...............(((((((.((((((.........(((((((((((..((....))(((.....))))).))))))))).))))))))))))). ( -47.00) >DroPse_CAF1 25691 120 - 1 CAGGCAGCCUUAUAAGCUGCCUGUUUGGUCUUCUCGUAGUCCUCUUUGGAGAUCUGGAGUUGCUGCCGGAGCAUUGCAAUCCGGCUUUGAGCGGCCUCGACCAGCUGCGUCUUGGAGGCC ((((((((.......))))))))...(((((((.((((((((.....)))...((((.(..(((((((((((...........)))))).)))))..)..)))))))))....))))))) ( -51.20) >DroEre_CAF1 1757 120 - 1 CAGGCUGCCUUGUAGGCGGCCUGCUUGGUCUUCUCAUAGUCGUCGUGGGCACUCACCAAAUGCUUUUGGAGCACGGCCAACCGGUUGCGAGCGGCUUCCAGGAGCUGCGACUUGGUGGCC (((((((((.....)))))))))...((((...(((.((((.(((..(.(.....(((((....))))).....((....))).)..)))(((((((....)))))))))))))).)))) ( -51.40) >DroWil_CAF1 44951 120 - 1 CAAGCUGCUUUGUAGGCGGCUUGUUUCGUCUUCUCAAAGUCUUCGCGAGCACUUUGCAAUUGCUUUUGGAGCAAACCUACACGGUUUUUGGCUGCCUCGAGUAAUUGAGUUUUCGAAGCC .((((((((.....))))))))((((((....(((((((((((...))).))))))(((((((((..((((((((((.....)))))...))).))..)))))))))))....)))))). ( -40.40) >DroMoj_CAF1 753 120 - 1 CAGGCCGCCCGAUAGGCCGCCUGCUUGGUCUUCUCAUAGUCAUCGCGUGCGUUGUCCAGCUGCUUUUGGAGCGUGGCCAGGCGGUUUUUGGCCGCCUCGAGCAGCUGCACCUUGGUGGUC ..(((((((.((((((((((.(((((((.....))).)).))..))).)).)))))(((((((((...(((.(((((((((.....)))))))))))))))))))))......))))))) ( -52.20) >DroPer_CAF1 25709 120 - 1 CAGGCAGCCUUAUAAGCUGCCUGUUUGGUCUUCUCGUAGUCCUCUUUGGAGAUCUGGAGUUGCUGCCGGAGCAUUGCAAUCCGGCUUUGAGCGGCCUCGAGCAGCUGCGUCUUGGAGGCC ((((((((.......))))))))...(((((((.((((((.(((....)))..((.((((((((((((((.........))))))....))))).))).))..))))))....))))))) ( -52.00) >consensus CAGGCAGCCUUAUAGGCGGCCUGCUUGGUCUUCUCAUAGUCCUCGUGGGCACUCUCCAGUUGCUUUUGGAGCAUGGCCAUCCGGUUUUGAGCGGCCUCGAGCAGCUGCGUCUUGGAGGCC (((((((((.....)))))))))...............((((((((....))....((((((((...((.((..((((....))))....))..))...))))))))......)))))). (-24.42 = -25.03 + 0.62)

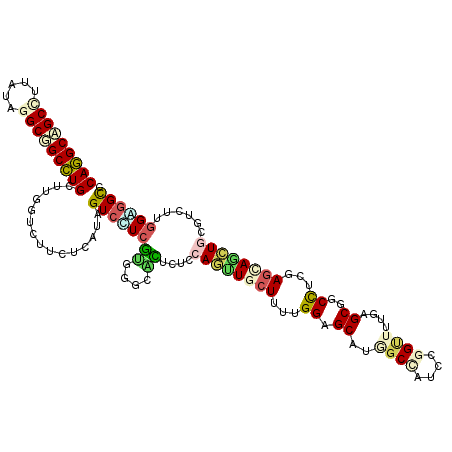
| Location | 12,341,297 – 12,341,408 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12341297 111 + 20766785 AUGGCCAUGCUCCAAAAGCAACUGGUGAGUGCCCACGACGACUAUGAGAAGACCAAGCAGGCCGCCUACAAGGCUGCCUGUGCCGCCGUGGAAGCCAAACAGAGGGCUGGA------ ..((((.((((.....)))).(((.((.((..(((((.((.((......)).....((((((.(((.....))).))))))..)).)))))..))))..)))..))))...------ ( -39.60) >DroPse_CAF1 25731 111 + 1 AUUGCAAUGCUCCGGCAGCAACUCCAGAUCUCCAAAGAGGACUACGAGAAGACCAAACAGGCAGCUUAUAAGGCUGCCUGUGCCGCCAUCGAGGCCAAGCAGAAGGCCGCC------ ...((........(((.((..(((.((.((((....)))).))..)))........((((((((((.....)))))))))))).))).....((((........)))))).------ ( -42.00) >DroWil_CAF1 44991 111 + 1 GUAGGUUUGCUCCAAAAGCAAUUGCAAAGUGCUCGCGAAGACUUUGAGAAGACGAAACAAGCCGCCUACAAAGCAGCUUGUGCUGCGGUUGAGGCCAAACAGAAGGCAUCA------ (((((((((((.....)))))...((((((.((.....)))))))).................))))))...(((((....)))))...(((.(((........))).)))------ ( -31.60) >DroYak_CAF1 661 111 + 1 UUGGCCUUGCUCCAAAAGCAACUGGUGAGUGCCCACGACGACUAUGAGAAGACCAAGCAGGCCGCCUACAAGGCAGCCUGUGCCGCCGUGGAGGCCAAACAGAGGGCUGGA------ (..((((((((.....)))..(((.((.((..(((((.((.((......)).....((((((.(((.....))).))))))..)).)))))..))))..))))))))..).------ ( -41.10) >DroMoj_CAF1 793 117 + 1 CUGGCCACGCUCCAAAAGCAGCUGGACAACGCACGCGAUGACUAUGAGAAGACCAAGCAGGCGGCCUAUCGGGCGGCCUGCGCUGCCGUCGAGGCCAAGCAGAAGGCGGCCAACAGC .(((((...(((.....((((((((.((.((....)).)).((......)).))).((((((.(((.....))).)))))))))))....)))(((........))))))))..... ( -49.10) >DroPer_CAF1 25749 111 + 1 AUUGCAAUGCUCCGGCAGCAACUCCAGAUCUCCAAAGAGGACUACGAGAAGACCAAACAGGCAGCUUAUAAGGCUGCCUGUGCCGCCAUCGAGGCCAAGCAGAAAGCCGCC------ ...((..((((..(((.....(((.((.((((....)))).))..)))........((((((((((.....)))))))))))))(((.....)))..))))....))....------ ( -37.80) >consensus AUGGCCAUGCUCCAAAAGCAACUGCAGAGCGCCCACGACGACUAUGAGAAGACCAAACAGGCCGCCUACAAGGCUGCCUGUGCCGCCGUCGAGGCCAAACAGAAGGCCGCA______ .((((..((((.....))))....................................((((((.(((.....))).)))))))))).......((((........))))......... (-22.92 = -23.03 + 0.12)
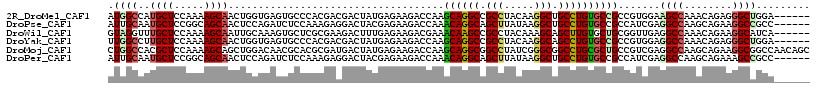
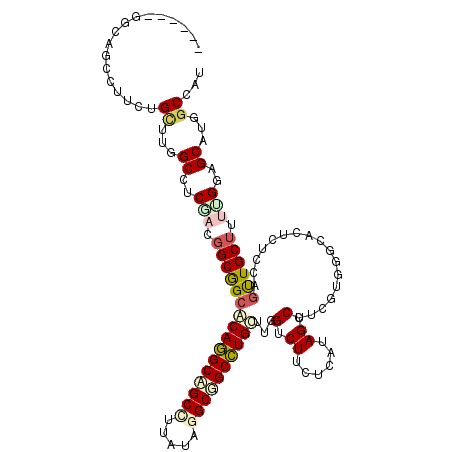
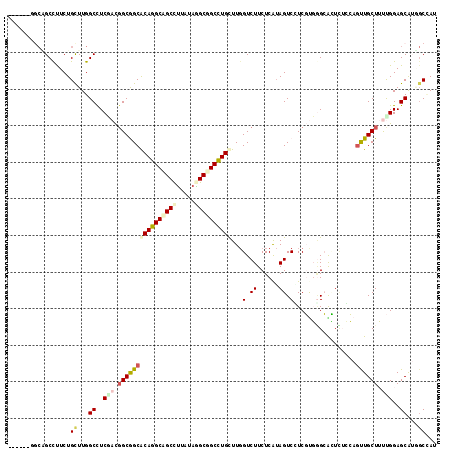
| Location | 12,341,297 – 12,341,408 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.42 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12341297 111 - 20766785 ------UCCAGCCCUCUGUUUGGCUUCCACGGCGGCACAGGCAGCCUUGUAGGCGGCCUGCUUGGUCUUCUCAUAGUCGUCGUGGGCACUCACCAGUUGCUUUUGGAGCAUGGCCAU ------..............((((((((((((((((.(((((.(((.....))).)))))..(((.....)))..)))))))))))......((((......)))).....))))). ( -41.90) >DroPse_CAF1 25731 111 - 1 ------GGCGGCCUUCUGCUUGGCCUCGAUGGCGGCACAGGCAGCCUUAUAAGCUGCCUGUUUGGUCUUCUCGUAGUCCUCUUUGGAGAUCUGGAGUUGCUGCCGGAGCAUUGCAAU ------.((((((........))))....((((((((((((((((.......)))))))))..(((((((....((....))..))))))).......)))))))..))........ ( -45.80) >DroWil_CAF1 44991 111 - 1 ------UGAUGCCUUCUGUUUGGCCUCAACCGCAGCACAAGCUGCUUUGUAGGCGGCUUGUUUCGUCUUCUCAAAGUCUUCGCGAGCACUUUGCAAUUGCUUUUGGAGCAAACCUAC ------(((.(((........))).)))...(((((....)))))...(((((..((((((...(.(((....))).)...)))))).........((((((...)))))).))))) ( -32.60) >DroYak_CAF1 661 111 - 1 ------UCCAGCCCUCUGUUUGGCCUCCACGGCGGCACAGGCUGCCUUGUAGGCGGCCUGCUUGGUCUUCUCAUAGUCGUCGUGGGCACUCACCAGUUGCUUUUGGAGCAAGGCCAA ------.............(((((((((((((((((.(((((((((.....)))))))))..(((.....)))..)))))))))))..........((((((...)))))))))))) ( -49.60) >DroMoj_CAF1 793 117 - 1 GCUGUUGGCCGCCUUCUGCUUGGCCUCGACGGCAGCGCAGGCCGCCCGAUAGGCCGCCUGCUUGGUCUUCUCAUAGUCAUCGCGUGCGUUGUCCAGCUGCUUUUGGAGCGUGGCCAG .....((((((((........)((..(((.((((((((((((.(((.....))).)))))).(((.(..(.(((.((....))))).)..).))))))))).)))..))))))))). ( -50.60) >DroPer_CAF1 25749 111 - 1 ------GGCGGCUUUCUGCUUGGCCUCGAUGGCGGCACAGGCAGCCUUAUAAGCUGCCUGUUUGGUCUUCUCGUAGUCCUCUUUGGAGAUCUGGAGUUGCUGCCGGAGCAUUGCAAU ------((((((.(((((((..(((.....))))))(((((((((.......)))))))))..(((((((....((....))..))))))).))))..))))))............. ( -43.60) >consensus ______GGCAGCCUUCUGCUUGGCCUCGACGGCGGCACAGGCAGCCUUAUAGGCGGCCUGCUUGGUCUUCUCAUAGUCCUCGUGGGCACUCUCCAGUUGCUUUUGGAGCAUGGCCAU .................((...((..(((.((((((((((((((((.....))))))))))...(.((......)).).................)))))).)))..))...))... (-27.72 = -28.42 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:58 2006