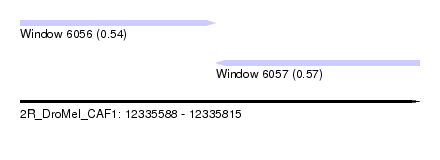
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,335,588 – 12,335,815 |
| Length | 227 |
| Max. P | 0.566784 |

| Location | 12,335,588 – 12,335,699 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.76 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -15.87 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
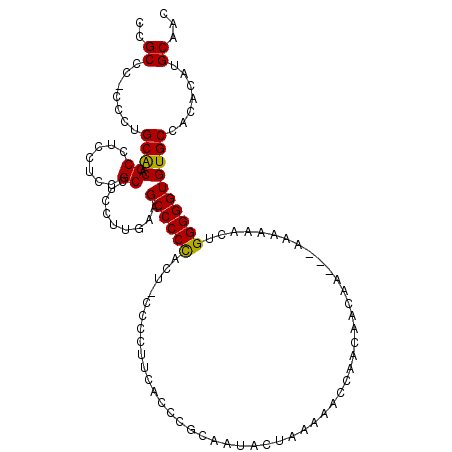
>2R_DroMel_CAF1 12335588 111 + 20766785 CCGCCC-CCCUGCACAGCCUCCUCCGCCUCCUUGAAGCCCCUACA-CCCCUUCAACUGCAAUAAUAAAAACCAACAAAAAAAAAAAAAACUGGGGGUGUGCCACACAUGCAAC .(((((-((..((...)).......((....((((((........-...))))))..))................................)))))))(((.......))).. ( -17.80) >DroSec_CAF1 34239 109 + 1 CCGCCCACCCGGCACAGCCUCCUCCGCCUCCUUGAAGCCCCCACU-CCCCUGCACCCGCAAUACUAAAAACCAACAACAA---CAAAAACUGGGGGUGUGCCACACAUGCAAC ..((......((((((.(((((...((.((...)).)).......-....(((....)))....................---........)))))))))))......))... ( -22.90) >DroEre_CAF1 34343 109 + 1 CCGCCC-CCCUGCGCAGCCUCCUCCGCCUCCUAGAAGCCCCCUCUUCCCCUUCACCCGCAAUACUGA-AACCAACAACAA--CAAAAAACCGGGGGUGUGCCACACAUGCAAC .(((((-((..(((.((....)).)))......((((......))))....................-............--.........)))))))(((.......))).. ( -19.60) >consensus CCGCCC_CCCUGCACAGCCUCCUCCGCCUCCUUGAAGCCCCCACU_CCCCUUCACCCGCAAUACUAAAAACCAACAACAA___AAAAAACUGGGGGUGUGCCACACAUGCAAC ..((.......((((.((.......)).........((((((.................................................)))))))))).......))... (-15.87 = -15.43 + -0.44)


| Location | 12,335,699 – 12,335,815 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
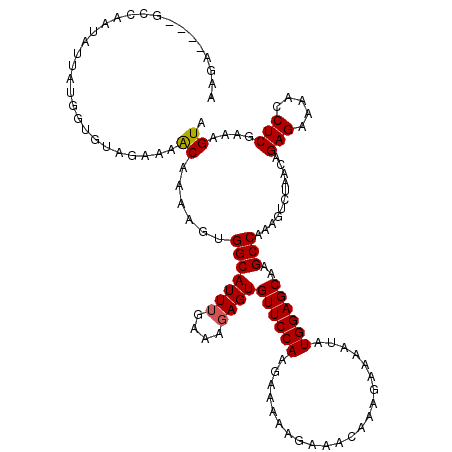
>2R_DroMel_CAF1 12335699 116 - 20766785 AGAA----GCCAAUAAUUUUGUGUAGAAAGCAAAAGUGGCAUUUGAAAGAGUGUUCCAAGAAAAAGAAACAAAGAAUAUAUGGAGCAAGCCAAAGUCUAACAGAGAAAACCUCGAAAGUA (((.----((((....((((((.......)))))).)))).((((...(..(((((((......................)))))))..))))).)))....(((.....)))....... ( -21.05) >DroSec_CAF1 34348 116 - 1 AGGA----GCUAAUAUUAUGUUGUAGAAAACAAAAGUGGCAUUUGAAAGAGUGUUCCAAGAAAAAGAAACAAACAAAAUAUGGAGCAAGCCAAAGUCUAACAGAGAAAACCUCGAAAGUA .((.----(((.(((((.((((((.....))....(..((((((....))))))..)..............)))).)))))..)))...))...........(((.....)))....... ( -20.00) >DroEre_CAF1 34452 120 - 1 AAGAGUUUGUUAAUAUUAGUGUGUUGAAAACAGAAGAGGCAUUAGAAAGAGUGUUCCAUGAAAAAGAAACAAAGAAAAUAUGGAGCAAGCCAAAGUCUAACAGAGAAAACCUCGAAAGUA .(((.((((......(((((((.((..........)).)))))))...(..(((((((((..................)))))))))..))))).)))....(((.....)))....... ( -22.27) >DroYak_CAF1 33893 115 - 1 AAC-----UUCAAUAUUAUGGUGUAGAAAACAGAAGUGGCAUUUGGAAGAGUGUUCCAAGAAAAAGAAACAAAGCAAAUAUGGAGCAAGCCAAAGUCUAACAGAGAAAACCUCGAAAGUA .((-----(((....................(((..((((.(((((((.....))))))).............((.........))..))))...)))....(((.....)))).)))). ( -19.40) >consensus AAGA____GCCAAUAUUAUGGUGUAGAAAACAAAAGUGGCAUUUGAAAGAGUGUUCCAAGAAAAAGAAACAAAGAAAAUAUGGAGCAAGCCAAAGUCUAACAGAGAAAACCUCGAAAGUA .............................((......(((((((....))))((((((......................))))))..)))...........(((.....)))....)). (-13.09 = -13.15 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:54 2006