
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,332,011 – 12,332,125 |
| Length | 114 |
| Max. P | 0.804480 |

| Location | 12,332,011 – 12,332,125 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -22.81 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12332011 114 + 20766785 AUAACGACAAUGCCACUAUCAACUGUGCUGCCAGAUAUAUGGGAGCCUGGUGGUACAGAGAAUGCCUUAGCAGGUAAGGUUUUUGACCUUUUCCCAAGAUUCCAAG------AUCAUAGU .......(((.(((........((((((..((((............))))..))))))....(((((....))))).)))..)))............((((....)------)))..... ( -28.50) >DroSec_CAF1 30705 109 + 1 AUAACGACAAUGCCACUAUCAACUGUGCCGCCAGAUACAUGGGAGCCUGGUGGUACAGGGAAUGUCUUAGCAGGUAA---UAUUGCCCAUUUCCG--CAUCCCAAG------AUCGUAGU ...(((((..............((((((((((((............))))))))))))((.((((.......((((.---...)))).......)--))).))..)------.))))... ( -31.94) >DroSim_CAF1 29156 112 + 1 AUAACGACAAUGCCACCAUCAACUGUGCCGCCAGAUACAUGGGAGCCUGGUGGUACAGAGAAUGUCUUAGCAGGUACGGUUUUUGCCCUUGCCCC--GAUCCCAAG------AUCGUAGU .....(((((((....)))...((((((((((((............))))))))))))....))))...(((((...(((....))))))))..(--((((....)------)))).... ( -35.00) >DroEre_CAF1 30627 112 + 1 ACAACGACAAUGCCACUAUCAACUGUGCCGCCAGAUACAUAGGAGCCUGGUGGUACAGGGAAUGUCUUGGCAGGUAAGGUUUUUGCCCUUUUCCG--GAGCCCAGA------ACCAACAU ..........(((((....((.((((((((((((............))))))))))))....))...))))).....((((((.((((......)--).))..)))------)))..... ( -36.80) >DroYak_CAF1 30176 112 + 1 AUAACGACAAUGCCACCAUCAAUUGUGCCGCCAGAUACAUGGGAGCUUGGUGGUACAGGGAAUGUCUGGGCAGGUAAGGAUUUUACCCUUUUUUG--AAUCCCAGG------AUCACCAU ..........((((((((........((..(((......)))..)).))))))))..((((...((((((((((.((((.......)))).))))--...))))))------.)).)).. ( -32.30) >DroAna_CAF1 28181 108 + 1 AUAACGAUCGUAGUAACAUUAAUUGUGCCGCCAAAUAUUUUGGGGCCUGGUGGUACAACGACUGCAUGGCCAGGUAAUUAUU----------CAA--GACUACAAGUUGUGGAACUUUUU (((((....(((((...........((((.((((.....))))((((..(..((......))..)..)))).))))......----------...--.)))))..))))).......... ( -27.40) >consensus AUAACGACAAUGCCACCAUCAACUGUGCCGCCAGAUACAUGGGAGCCUGGUGGUACAGGGAAUGUCUUAGCAGGUAAGGUUUUUGCCCUUUUCCG__GAUCCCAAG______AUCAUAGU ..........((((........((((((((((((............))))))))))))....(((....)))))))............................................ (-22.81 = -22.32 + -0.49)

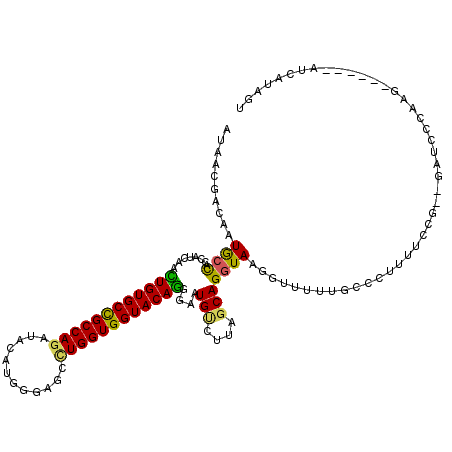
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:48 2006