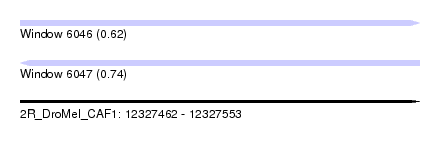
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,327,462 – 12,327,553 |
| Length | 91 |
| Max. P | 0.740987 |

| Location | 12,327,462 – 12,327,553 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -19.05 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12327462 91 + 20766785 -----------------------------AAAAAGGAGAAACAGGCAUGUUGAACAAGCGAUGUCGCCCGUCUUGUAAAACCUGCAACACAUGCAACAUGCAACUUGCAACUUGCAUCAG -----------------------------........((.....((((((((.....(((....)))..((.(((((.....))))).))...))))))))....(((.....))))).. ( -21.80) >DroSec_CAF1 26188 91 + 1 -----------------------------AAAAAGGAGAAACAGGCAUGUUGAACAAGUGAUGUCGCUCGUCUUGUAAAACCUGCAACACAUGCAACAUGCAACUUGCAACUUGCAUCAG -----------------------------...............(((.((((..(((((.((((.((..((.(((((.....))))).))..)).))))...))))))))).)))..... ( -21.60) >DroSim_CAF1 24370 91 + 1 -----------------------------AAAAAGGAGAAACAGGCAUGUUGAACAAGCGAUGUCGCCCGUCUUGUAAAACCUGCAACACAUGCAACAUGCAACUAGCAACUUGCAUCAG -----------------------------........((.....((((((((.....(((....)))..((.(((((.....))))).))...)))))))).....((.....)).)).. ( -21.30) >DroEre_CAF1 26456 91 + 1 -----------------------------GAAAAGGAGGUACAGGCAUGUUGAACAAGCGAUGUCGCCCGUCUUGUAAAACCUGCAACACAUGCAACAUGUAACUUGCAACUUGCAUCAG -----------------------------((..(((..(((...((((((((.....(((....)))..((.(((((.....))))).))...))))))))....)))..)))...)).. ( -21.60) >DroYak_CAF1 25573 91 + 1 -----------------------------GAAAAGGAGGCACAGGCAUGUUGAACAAGCGAUGUCGCCCGUCUUGUAAAACCUGCAACACAUGCAACAUGUAACUUGCAACUUGCAUCAG -----------------------------((..(((..(((...((((((((.....(((....)))..((.(((((.....))))).))...))))))))....)))..)))...)).. ( -23.60) >DroAna_CAF1 24008 119 + 1 AAAUAUACAACACAUAUAUACCUACAAAUAAAAAGGAGAAACAGGCAUGUUGAACAAGCCUUGUCGCUCGUCUUGUAAAACCUGCAACACAUGCAACAUGCAACUUGCAACUCGUC-CGG .....(((((.((...(((........))).....(((..((((((.((.....)).))).)))..))))).))))).....(((((....(((.....)))..))))).......-... ( -21.60) >consensus _____________________________AAAAAGGAGAAACAGGCAUGUUGAACAAGCGAUGUCGCCCGUCUUGUAAAACCUGCAACACAUGCAACAUGCAACUUGCAACUUGCAUCAG ............................................((((((((.....(((....)))..((.(((((.....))))).))...)))))))).....((.....))..... (-19.05 = -18.72 + -0.33)

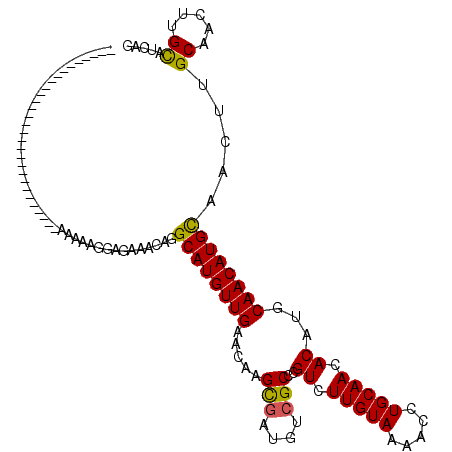
| Location | 12,327,462 – 12,327,553 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -23.53 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12327462 91 - 20766785 CUGAUGCAAGUUGCAAGUUGCAUGUUGCAUGUGUUGCAGGUUUUACAAGACGGGCGACAUCGCUUGUUCAACAUGCCUGUUUCUCCUUUUU----------------------------- ...((((((..(((.....)))..))))))..(..((((((.......((((((((....))))))))......))))))..)........----------------------------- ( -29.12) >DroSec_CAF1 26188 91 - 1 CUGAUGCAAGUUGCAAGUUGCAUGUUGCAUGUGUUGCAGGUUUUACAAGACGAGCGACAUCACUUGUUCAACAUGCCUGUUUCUCCUUUUU----------------------------- ...((((((..(((.....)))..))))))..(..((((((.......((((((.(....).))))))......))))))..)........----------------------------- ( -24.52) >DroSim_CAF1 24370 91 - 1 CUGAUGCAAGUUGCUAGUUGCAUGUUGCAUGUGUUGCAGGUUUUACAAGACGGGCGACAUCGCUUGUUCAACAUGCCUGUUUCUCCUUUUU----------------------------- ...((((((..(((.....)))..))))))..(..((((((.......((((((((....))))))))......))))))..)........----------------------------- ( -29.32) >DroEre_CAF1 26456 91 - 1 CUGAUGCAAGUUGCAAGUUACAUGUUGCAUGUGUUGCAGGUUUUACAAGACGGGCGACAUCGCUUGUUCAACAUGCCUGUACCUCCUUUUC----------------------------- .....(((...(((((........)))))..)))(((((((.......((((((((....))))))))......)))))))..........----------------------------- ( -25.12) >DroYak_CAF1 25573 91 - 1 CUGAUGCAAGUUGCAAGUUACAUGUUGCAUGUGUUGCAGGUUUUACAAGACGGGCGACAUCGCUUGUUCAACAUGCCUGUGCCUCCUUUUC----------------------------- ..((((((...(((((........)))))))))))((((((.......((((((((....))))))))......))))))...........----------------------------- ( -25.02) >DroAna_CAF1 24008 119 - 1 CCG-GACGAGUUGCAAGUUGCAUGUUGCAUGUGUUGCAGGUUUUACAAGACGAGCGACAAGGCUUGUUCAACAUGCCUGUUUCUCCUUUUUAUUUGUAGGUAUAUAUGUGUUGUAUAUUU ..(-((((((((((.....)).((((((.(((.(((.........))).))).)))))).)))))))))...(((((((.................)))))))................. ( -28.43) >consensus CUGAUGCAAGUUGCAAGUUGCAUGUUGCAUGUGUUGCAGGUUUUACAAGACGGGCGACAUCGCUUGUUCAACAUGCCUGUUUCUCCUUUUU_____________________________ ...((((((..(((.....)))..))))))..(..((((((.......((((((((....))))))))......))))))..)..................................... (-23.53 = -24.34 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:44 2006