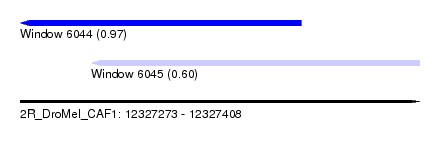
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,327,273 – 12,327,408 |
| Length | 135 |
| Max. P | 0.969563 |

| Location | 12,327,273 – 12,327,368 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12327273 95 - 20766785 AAAUGAACA---------AAAAACACAAACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAGUACCAACUUAUGAAGUCCGGGUACAGUGGUACUUCG------ .........---------..............((.(((((((((((....)))))))..........(((((.(((.....)))...)))))))))))......------ ( -19.70) >DroSec_CAF1 25993 101 - 1 AAAUGGAGAAAAAAACAA-AAA--ACACACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAUUACCAACUUAUGAAGUCCGGGUACAGUGGUAAUUCG------ ..................-...--........((.(((((((((((....)))))))...........((((.(((.....)))...)))).))))))......------ ( -15.80) >DroSim_CAF1 24172 104 - 1 AAAUGAAGAAAAAAACCAAAAAACACACACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAGUACCAACUUAUGAAGUCCGGGUACAGUGGUACUUCG------ ....((((......((((....................((((((((....)))))))).........(((((.(((.....)))...)))))..)))).)))).------ ( -20.90) >DroEre_CAF1 26267 91 - 1 AAAUGAAAAAA------C-AA------AACACACACACUGCCACAGCAACUUGUUGCACAAAUA------CCAAGGAAUGAAGUCCGGGUACAGUGGUGCUUCGAGCGAC ...........------.-..------....(((.(((((.....((((....)))).....((------((..(((......))).))))))))))))........... ( -19.70) >DroYak_CAF1 25391 90 - 1 AAAUGAAAAAA---AUAA-AA------AACACACACACUGCGACAGCAACUUGUUGCACAAAUA------CCAGCGAAUGGAGUCCGGGUACAGUGGUACUUCG----AC ...........---....-..------...........((((((((....))))))))......------(((.....))).(((..(((((....)))))..)----)) ( -18.30) >consensus AAAUGAAAAAA___A__A_AAA__ACAAACACACACACUGCAACAGCAACUUGUUGCACAAAUACAA_UACCAACUUAUGAAGUCCGGGUACAGUGGUACUUCG______ ......................................((((((((....)))))))).....................((((((((.......))).)))))....... (-13.40 = -13.36 + -0.04)

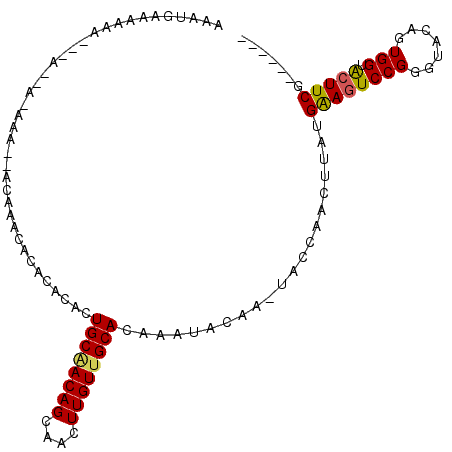
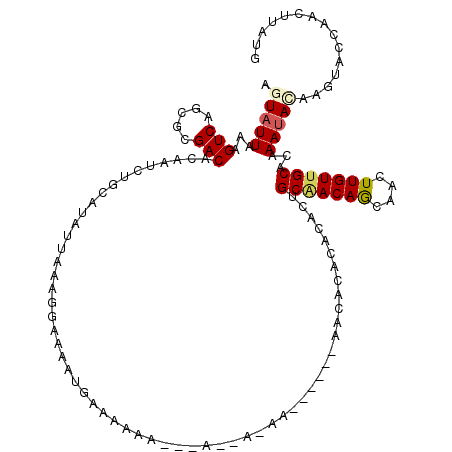
| Location | 12,327,297 – 12,327,408 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -14.78 |
| Consensus MFE | -10.06 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12327297 111 - 20766785 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGAACA---------AAAAACACAAACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAGUACCAACUUAUG .(((((...(((.....))).............................---------....................((((((((....))))))))..)))))((((....))))... ( -14.40) >DroSec_CAF1 26017 117 - 1 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGGAGAAAAAAACAA-AAA--ACACACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAUUACCAACUUAUG .(((((...(((.....)))....(((.(((.((.....)).))).))).........-...--..............((((((((....))))))))..)))))............... ( -16.90) >DroSim_CAF1 24196 120 - 1 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGAAGAAAAAAACCAAAAAACACACACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAGUACCAACUUAUG .(((((...(((.....)))....(((.(((.((.....)).))).))).............................((((((((....))))))))..)))))((((....))))... ( -15.40) >DroEre_CAF1 26297 101 - 1 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGAAAAAA------C-AA------AACACACACACUGCCACAGCAACUUGUUGCACAAAUA------CCAAGGAAUG .(((((...(....)((((((...(((..........)))...........------.-..------...........(((....)))...))))))...))))------)......... ( -9.60) >DroYak_CAF1 25417 104 - 1 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGAAAAAA---AUAA-AA------AACACACACACUGCGACAGCAACUUGUUGCACAAAUA------CCAGCGAAUG .(((((...(((.....)))...............................---....-..------...........((((((((....))))))))..))))------)......... ( -12.60) >DroAna_CAF1 23840 95 - 1 AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAAGAAAACGAAAAAA---A----------------GAGAUGUGGCAACAACAACUUGUUGCACAAACAUCAGUAC------AGG .(((((...(((.....)))...............................---.----------------..(((((.(((((((....)))))))....))))))))))------... ( -19.80) >consensus AGUAUUAAAGUCAGCGCGACACAAUCUGCAUAUUAAAGGAAAAUGAAAAAA___A__A_AA______AACACACACACUGCAACAGCAACUUGUUGCACAAAUACAAGUACCAACUUAUG .(((((...(((.....)))...........................................................(((((((....)))))))...)))))............... (-10.06 = -10.40 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:42 2006