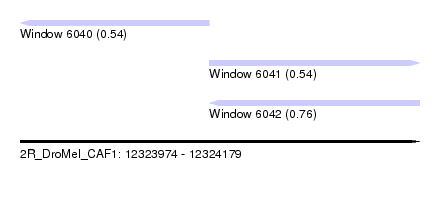
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,323,974 – 12,324,179 |
| Length | 205 |
| Max. P | 0.761228 |

| Location | 12,323,974 – 12,324,071 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -20.03 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12323974 97 - 20766785 ACAAAUUUCAAUUCCACACCGCGUGAGUGUGCGUUAUAAAUUGCGUAUGCAGGCCCACUUGGUCAGUUU-----------------------GUGAAUACAAAUCUACGCGUUUUACAUA ...................((((((.((((((((........)))))))).((((.....)))).((((-----------------------((....)))))).))))))......... ( -23.20) >DroSec_CAF1 22760 97 - 1 ACAAAUUUCAAUUCCACACCGCGUGAGUGUGCGUUAUAAAUUGCGUAUGCAGGCCCACUUGGUCAGUUU-----------------------GUGAAUACAAAUCUACGCGUUUUACAUA ...................((((((.((((((((........)))))))).((((.....)))).((((-----------------------((....)))))).))))))......... ( -23.20) >DroSim_CAF1 20920 97 - 1 ACAAAUUUCAAUUCCACACCGCGUGAGUGUGCGUUAUAAAUUGCGUAUGCAGGCCCACUUGGUCAGUUU-----------------------GUGAAUACAAAUCUACGCGUUUUACAUA ...................((((((.((((((((........)))))))).((((.....)))).((((-----------------------((....)))))).))))))......... ( -23.20) >DroEre_CAF1 22985 97 - 1 ACAAAUUUCAAUUCCACACCGCGUAAGUGUGCGUUAUAAAUUGCGUAUGCAGACCCACUUGGUCAGUUU-----------------------GUGAAUACAAAUCUACGCGUUUUACAUA ...................((((((.((((((((........)))))))).((((.....)))).((((-----------------------((....)))))).))))))......... ( -24.10) >DroYak_CAF1 21360 97 - 1 ACAAAUUUCAAUUCCACACCGCGUGAGUGUGCGUUAUAAAUUGCGUAUGCAGGGCCACUUGGUCAGUUU-----------------------GUGAAUACAAAUCUACGCGCUUUACAUA ....................(((((.((((((((........))))))))..((((....)))).((((-----------------------((....))))))...)))))........ ( -24.10) >DroAna_CAF1 20512 120 - 1 ACAAAUUUCAAUUCCAUUCCGUAUGUGUGUGCGUUAUAAAUUGCGUAUGCAGACCCAUUUGGUCAGUUUUUUUUUUGAUUUUUUUUUAGGUUUUGAAUACAAAUCUACGCGUUUUACAUA ......(((((..((..........(((((((((........)))))))))((((.....))))........................))..)))))....................... ( -22.00) >consensus ACAAAUUUCAAUUCCACACCGCGUGAGUGUGCGUUAUAAAUUGCGUAUGCAGGCCCACUUGGUCAGUUU_______________________GUGAAUACAAAUCUACGCGUUUUACAUA ...................((((((.((((((((........)))))))).((((.....)))).........................................))))))......... (-20.03 = -19.87 + -0.16)

| Location | 12,324,071 – 12,324,179 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12324071 108 + 20766785 CUCAGUCCAGGUCGGUCUCCCGGGUUUCAGGUCCCCAAAGACCCAAAAG-----ACCCGCGUCCACCGACCGGCGCUUCUUGCUUUUCAAUCGACUAAAAGCUAUGAAACCGA ....((((.(((((((..(.(((((((..((((......))))....))-----))))).)...))))))))).))(((..((((((.........))))))...)))..... ( -34.00) >DroSec_CAF1 22857 94 + 1 CUCAGUCCAGGUCGGUCCCCCGGGUUUCAGGUCCCCAAAGACCCAA-AG-----ACC-------------CGGCGCUUCUUGCUUUUCAAUCGACUAAAAGCUAUGAAACCGA ...........(((((...((((((((..((((......))))...-))-----)))-------------)))...(((..((((((.........))))))...)))))))) ( -27.50) >DroSim_CAF1 21017 94 + 1 CUCAGUCCAGGUCGGUCCCCCGGGUUUCAGGUCCCCAAAGACCCAA-AG-----ACC-------------CGGCGCUUCUUGCUUUUCAAUCGACUAAAAGCUAUGAAACCGA ...........(((((...((((((((..((((......))))...-))-----)))-------------)))...(((..((((((.........))))))...)))))))) ( -27.50) >DroEre_CAF1 23082 85 + 1 CUCAGUCCAGGUCGGUCCCCCGGGUUUCAGGUCCCCAAAGACC----------------CGUCCACCGCCCGGCGCUUCUUGCUUUUCAAUCGA------------AAACCGA ...(((((.((.((((....(((((((..((...))..)))))----------------))...)))))).)).)))..(((.(((((....))------------))).))) ( -23.80) >DroYak_CAF1 21457 113 + 1 CUCAGUCCAGGUCGGUUCCCUGGGUUUCAGGUCCCCAAAGACCCCCGUCCCCUUUCCACCGUCCACCACCCGGCGCUUCUUGCUUUUCAAUCGACUAAAAGCUAUGAAGCCGA .........((.((((.....(((..(..((((......))))...)..))).....)))).))......((((...((..((((((.........))))))...)).)))). ( -30.10) >consensus CUCAGUCCAGGUCGGUCCCCCGGGUUUCAGGUCCCCAAAGACCCAA_AG_____ACC__CGUCCACC__CCGGCGCUUCUUGCUUUUCAAUCGACUAAAAGCUAUGAAACCGA ............(((....)))(((((((((((......))))............................((((.....))))....................))))))).. (-16.10 = -16.18 + 0.08)

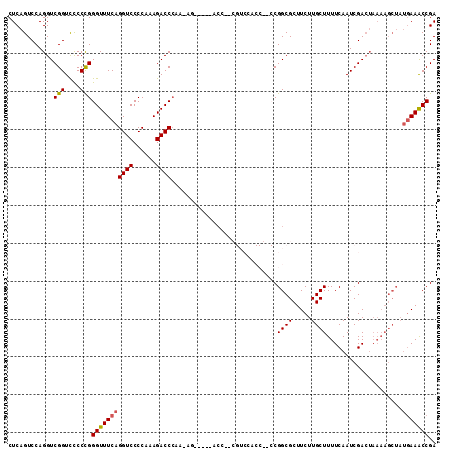
| Location | 12,324,071 – 12,324,179 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
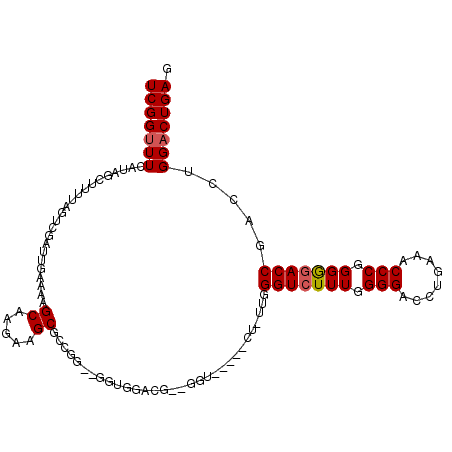
>2R_DroMel_CAF1 12324071 108 - 20766785 UCGGUUUCAUAGCUUUUAGUCGAUUGAAAAGCAAGAAGCGCCGGUCGGUGGACGCGGGU-----CUUUUGGGUCUUUGGGGACCUGAAACCCGGGAGACCGACCUGGACUGAG (((((......((((((.((..........)).)))))).(((((((((...(.(((((-----..((..(((((....)))))..))))))).)..))))))).))))))). ( -43.60) >DroSec_CAF1 22857 94 - 1 UCGGUUUCAUAGCUUUUAGUCGAUUGAAAAGCAAGAAGCGCCG-------------GGU-----CU-UUGGGUCUUUGGGGACCUGAAACCCGGGGGACCGACCUGGACUGAG (((((..(...((((((.((..........)).)))))).(((-------------(((-----.(-(..(((((....)))))..)))))))))..)))))........... ( -38.70) >DroSim_CAF1 21017 94 - 1 UCGGUUUCAUAGCUUUUAGUCGAUUGAAAAGCAAGAAGCGCCG-------------GGU-----CU-UUGGGUCUUUGGGGACCUGAAACCCGGGGGACCGACCUGGACUGAG (((((..(...((((((.((..........)).)))))).(((-------------(((-----.(-(..(((((....)))))..)))))))))..)))))........... ( -38.70) >DroEre_CAF1 23082 85 - 1 UCGGUUU------------UCGAUUGAAAAGCAAGAAGCGCCGGGCGGUGGACG----------------GGUCUUUGGGGACCUGAAACCCGGGGGACCGACCUGGACUGAG (((((..------------...........((.....)).(((((.(((...((----------------(((((....)))))))..)))(((....))).)))))))))). ( -32.80) >DroYak_CAF1 21457 113 - 1 UCGGCUUCAUAGCUUUUAGUCGAUUGAAAAGCAAGAAGCGCCGGGUGGUGGACGGUGGAAAGGGGACGGGGGUCUUUGGGGACCUGAAACCCAGGGAACCGACCUGGACUGAG ...(((((...((((((.........))))))..))))).(((((((((...(........(....)(((..((...(....)..))..))).)...))).))))))...... ( -36.80) >consensus UCGGUUUCAUAGCUUUUAGUCGAUUGAAAAGCAAGAAGCGCCGG__GGUGGACG__GGU_____CU_UUGGGUCUUUGGGGACCUGAAACCCGGGGGACCGACCUGGACUGAG (((((((.......................((.....))...............................(((((((.(((........))).))))))).....))))))). (-20.28 = -20.52 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:39 2006