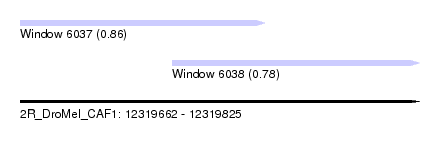
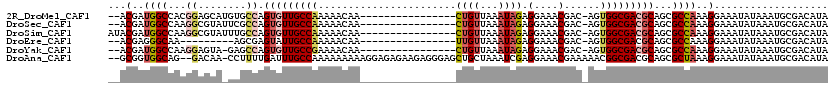
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,319,662 – 12,319,825 |
| Length | 163 |
| Max. P | 0.862956 |

| Location | 12,319,662 – 12,319,762 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
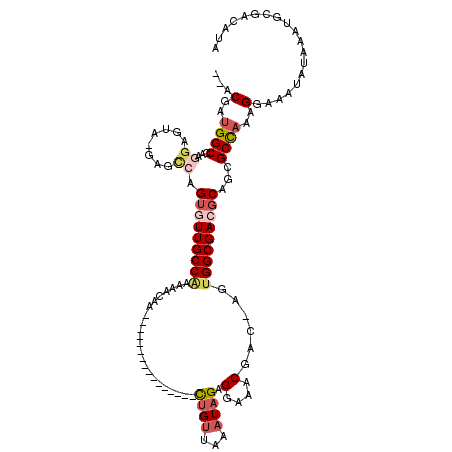
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
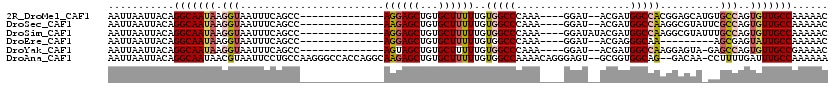
>2R_DroMel_CAF1 12319662 100 + 20766785 AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC--------------AGGAGCUGUGCUUUUUGUGGCCCAAA----GGAU--ACGAUGGCCACGGAGCAUGUGCCAGUGUUGCCAAAAAC ...........(((((((.((((....((((.--------------....))))(((((..(((((((....----(...--.)...))))))))))))..))))..)))))))...... ( -34.30) >DroSec_CAF1 18444 100 + 1 AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC--------------AAGAGCUGUGCUUUUUGUGGCCCAAA----GGAU--ACGAUGGCCAAGGCGUAUUCGCCAGUGUUGCCAAAAAC ...........((((((((((((....((((.--------------....)))))))))....(((((....----(...--.)...))))).((((....))))..)))))))...... ( -29.20) >DroSim_CAF1 16551 102 + 1 AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC--------------AGGAGCUGUGCUUUUUGUGGCCCAAA----GGAUAUACGAUGGCCAAGGCGUAUUUGCCAGUGUUGCCAAAAAC ...........(((((((.(((((.....(((--------------((((((...))))))..(((((....----(......)...))))).)))....)))))..)))))))...... ( -31.60) >DroEre_CAF1 18486 91 + 1 AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC--------------AGGAGCUGUGCUUUUUGUGGCCCAAA----GGAU--ACGAGGGCAA---------AGCGAGUAUUGCCAAAAAC ...........(((((((.(((.......)))--------------....(((.(((((((.(((.((....----)).)--))))))))).---------)))...)))))))...... ( -26.30) >DroYak_CAF1 16851 99 + 1 AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC--------------AGUAGCUGUGCUUUUUGUGGCCCAAA----GGAU--ACGAUGGCCAAGGAGUA-GAGCCAGUGUUGCCGAAAAC ...........(((((((.(((.......)))--------------....(((.((((((...(((((....----(...--.)...))))).))))))-.)))...)))))))...... ( -30.80) >DroAna_CAF1 16469 115 + 1 AAUUAAUUACAGGCAAUAACGUAAUUCCUGCCAAGGGCCACCAGGCAAGAGCUGUGCUUUUUGUGGCCAAAACAGGGAGU--GCGGUGGCAG--GACAA-CCUUUUGAUUUGCCAAAAAA ...........(((((.........(((((((....(((....)))....((((..((((((((.......)))))))).--.)))))))))--))(((-....)))..)))))...... ( -40.90) >consensus AAUUAAUUACAGGCAAUAAGGUAAUUUCAGCC______________AGGAGCUGUGCUUUUUGUGGCCCAAA____GGAU__ACGAUGGCCAAGGAGUA_GAGCCAGUGUUGCCAAAAAC ...........(((((((.(((........................((((((...))))))..(((((...................)))))..........)))..)))))))...... (-15.72 = -16.44 + 0.73)
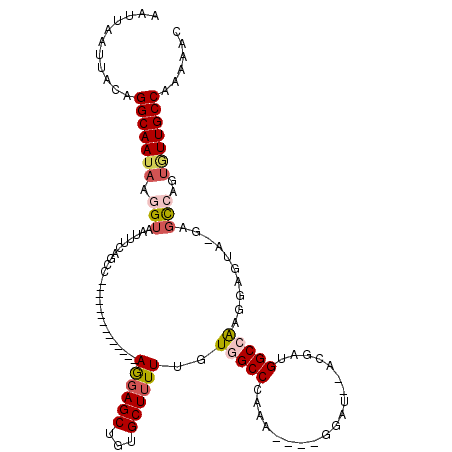
| Location | 12,319,724 – 12,319,825 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -16.36 |
| Energy contribution | -17.53 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12319724 101 + 20766785 --ACGAUGGCCACGGAGCAUGUGCCAGUGUUGCCAAAAACAA----------------CUGUUAAAUAGAGGAAACGAC-AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA --....((((.(((.....)))))))(((((((........(----------------(((((.......(....))))-)))((((......))))...............))))))). ( -26.91) >DroSec_CAF1 18506 101 + 1 --ACGAUGGCCAAGGCGUAUUCGCCAGUGUUGCCAAAAACAA----------------CUGUUAAAUAGAGGAAACGAC-AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA --.(..(((((..((((....)))).(((((((((.......----------------((((...)))).(....)...-..))))))))).).))))..)................... ( -30.30) >DroSim_CAF1 16613 103 + 1 AUACGAUGGCCAAGGCGUAUUUGCCAGUGUUGCCAAAAACAA----------------CUGUUAAAUAGAGGAAACGAC-AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA ...(..(((((..((((....)))).(((((((((.......----------------((((...)))).(....)...-..))))))))).).))))..)................... ( -28.90) >DroEre_CAF1 18548 92 + 1 --ACGAGGGCAA---------AGCGAGUAUUGCCAAAAACAA----------------UUGUUAAAUAGAGGAAACGAC-AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA --.(...(((..---------.((..(..((((((..(((..----------------..))).......(....)...-..))))))..).)))))...)................... ( -18.10) >DroYak_CAF1 16913 100 + 1 --ACGAUGGCCAAGGAGUA-GAGCCAGUGUUGCCGAAAACAA----------------CUGUUAAAUAGAGGAAACGAC-AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA --....(((((........-).))))(((((((........(----------------(((((.......(....))))-)))((((......))))...............))))))). ( -25.11) >DroAna_CAF1 16549 115 + 1 --GCGGUGGCAG--GACAA-CCUUUUGAUUUGCCAAAAAAAAAGGAGAGAAGAGGGAGCUGCUAAAUCGAGGAAACGAAAAACGGCGACGCAGCGCUAAAGGAAAUAUAAAUGCGACAUA --...((.(((.--.....-((((((..(((.((.........)))))..)))))).(((((........(....).......(....)))))).................))).))... ( -26.10) >consensus __ACGAUGGCCAAGGAGUA_GAGCCAGUGUUGCCAAAAACAA________________CUGUUAAAUAGAGGAAACGAC_AGUGGCGACGCAGCGCCAAAGGAAAUAUAAAUGCGACAUA ...(..((((...((........)).(((((((((.......................((((...)))).(....)......)))))))))...))))..)................... (-16.36 = -17.53 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:36 2006