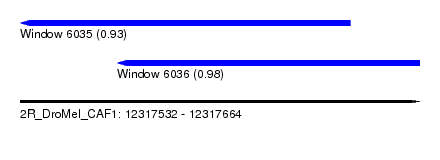
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,317,532 – 12,317,664 |
| Length | 132 |
| Max. P | 0.984739 |

| Location | 12,317,532 – 12,317,641 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
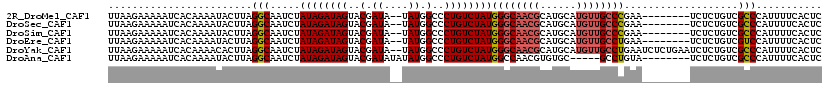
>2R_DroMel_CAF1 12317532 109 - 20766785 UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCCGAA--------UCUCUGUCGCCCAUUUUCACUC ........................(((.....((((((((..(....--....)..))))))))((((((((......))))))))...--------........)))........... ( -25.90) >DroSec_CAF1 16286 109 - 1 UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCCGAA--------UCUCUGUCGCCCAUUUUCACUC ........................(((.....((((((((..(....--....)..))))))))((((((((......))))))))...--------........)))........... ( -25.90) >DroSim_CAF1 14451 109 - 1 UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCCGAA--------UCUCUGUCGCCCAUUUUCACUC ........................(((.....((((((((..(....--....)..))))))))((((((((......))))))))...--------........)))........... ( -25.90) >DroEre_CAF1 16413 109 - 1 UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCUGAA--------UCUCUGUCGUCCAUUUUCACUC ....(((((((.(((......(((((((((((((((((((..(....--....)..))))))))))(((.....)))..))))))))).--------....))).))...))))).... ( -26.80) >DroYak_CAF1 14751 117 - 1 UUAAGAAAAAUCACAAAACACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCUGAAUCUCUGAAUCUCUGUCGCCCAUUUUCACUC ....(((((............(((((((((((((((((((..(....--....)..))))))))))(((.....)))..)))))))))......((.......)).....))))).... ( -26.40) >DroAna_CAF1 14003 106 - 1 UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUAUAUAUGGCCCUGUCUAUGGCCAACGUGUGC-----GCCUGUA--------UCUCUGUCGCCCAUUUUCACUC ....(((((...(((..((((..((((.(((....))).(((((.......(((((........)))))...)))))-----)))))))--------)...)))......))))).... ( -21.40) >consensus UUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA__UAUGGCCCUGUCUAUGGGCAACGCAUGCAUGUUGCCCGAA________UCUCUGUCGCCCAUUUUCACUC ........................(((.....((((((((..(.((....)).)..))))))))((((((((......))))))))...................)))........... (-22.52 = -22.97 + 0.44)
| Location | 12,317,564 – 12,317,664 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -20.00 |
| Consensus MFE | -18.49 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
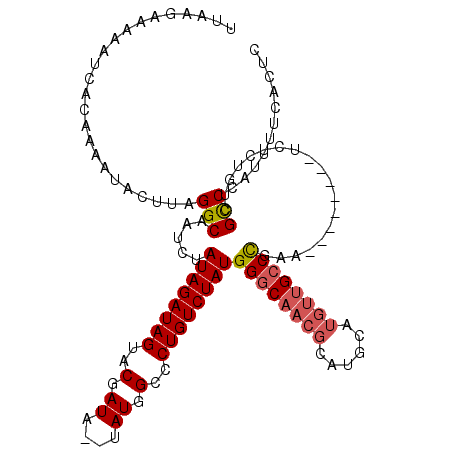
>2R_DroMel_CAF1 12317564 100 - 20766785 AAGGCAAC--ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((..--.........((.(((((......)))))))..........((..(((((((((((..(....--....)..)))))))))))....)).))).. ( -19.30) >DroSec_CAF1 16318 100 - 1 AAGGCAAC--ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((..--.........((.(((((......)))))))..........((..(((((((((((..(....--....)..)))))))))))....)).))).. ( -19.30) >DroSim_CAF1 14483 100 - 1 AAGGCAAC--ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((..--.........((.(((((......)))))))..........((..(((((((((((..(....--....)..)))))))))))....)).))).. ( -19.30) >DroEre_CAF1 16445 100 - 1 AAGGCAAC--ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((..--.........((.(((((......)))))))..........((..(((((((((((..(....--....)..)))))))))))....)).))).. ( -19.30) >DroYak_CAF1 14791 100 - 1 AAGGCAAC--ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAACACUUAGGCAAUCUAUAGAUAGUACGAUA--UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((..--.........((.(((((......)))))))..........((..(((((((((((..(....--....)..)))))))))))....)).))).. ( -19.30) >DroAna_CAF1 14032 102 - 1 GAACUAACACACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUAUAUAUGGCCCUGUCUAUGGCCAACGUGUGC-- .......(((((.......((.(((((......))))))).........(((....(((((((((..(.((....)).)..))))))))))))...))))).-- ( -23.50) >consensus AAGGCAAC__ACAAAUUUAUGAUGAUUAAGAAAAAUCACAAAAUACUUAGGCAAUCUAUAGAUAGUACGAUA__UAUGGCCCUGUCUAUGGGCAACGCAUGCAU ...(((.............((.(((((......)))))))..........((..(((((((((((..(.((....)).)..)))))))))))....)).))).. (-18.49 = -18.55 + 0.06)

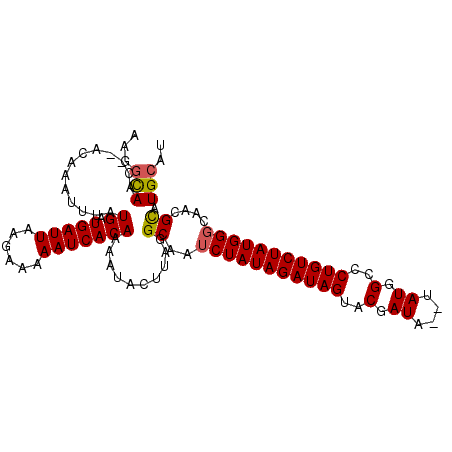
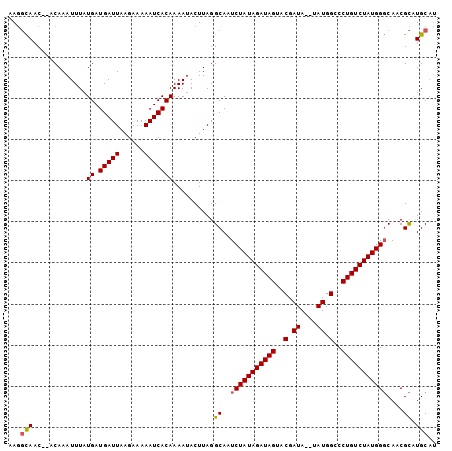
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:34 2006