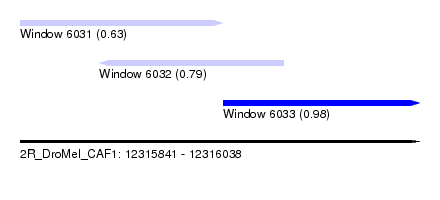
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,315,841 – 12,316,038 |
| Length | 197 |
| Max. P | 0.978184 |

| Location | 12,315,841 – 12,315,941 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -14.44 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
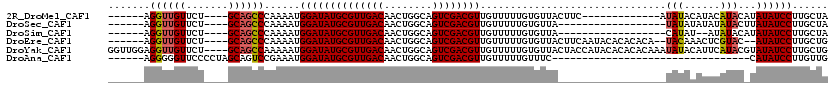
>2R_DroMel_CAF1 12315841 100 + 20766785 GGAAAACCAAAAAGAGG-GGACACUUAACUGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACAAACAGGAGUUGGGG------------UCCUCGAAAAGGA-----CUCCCUUCAG-- ((....)).....((((-(((.......(..((((((((((((......((((...))))))))))).)))))..).(------------((((.....))))-----))))))))..-- ( -36.00) >DroSec_CAF1 14609 100 + 1 GGAAAGCCAAAAAGAGG-GGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACAAACAGGAGUUGGGG------------UCCUCCAAAAGGA-----CCCCCUUCAG-- .(((.(((...(((.(.-...).)))....))))))(((((((......((((...))))))))))).((((..((((------------((((.....))))-----))))))))..-- ( -36.40) >DroSim_CAF1 12764 100 + 1 GGAAAGCCAAAAAGAGG-GGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACAAACAGGAGUUGGGG------------UCCUCCAAAAGGA-----CCCCCUUCAG-- .(((.(((...(((.(.-...).)))....))))))(((((((......((((...))))))))))).((((..((((------------((((.....))))-----))))))))..-- ( -36.40) >DroEre_CAF1 14766 97 + 1 -GAAAACCAAAAAGAGG-GGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACA-ACAGGAGUCGGGG------------UCCACCUAAGGGG-----GU-CCUUCAG-- -(((.(((......(((-((((.(((....((((((...(((((...((((((...))))))))-)))))))))))))------------))).))).....)-----))-..)))..-- ( -30.20) >DroYak_CAF1 13053 87 + 1 GGAAAACCAAAAAGAGG-GGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACA-ACAGGAGUUGGG-------------------------G-----GU-CCUUCAGAG ((....)).....((((-((...(((....((((((...(((((...((((((...))))))))-))))))))).))-------------------------)-----.)-))))).... ( -23.90) >DroAna_CAF1 12356 117 + 1 GGAUGGAGAGGGAGAGGUCGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCUAAACCAAACAAA-AGGAGGUGGGGGUGUCCUUUUCCUCCGCCGAAAGGAGGAGGGUCCUUUCCA-- ...((((((((((((((...((((((....(.((((.((((((......(((.....)))))))))-.)))).)..))))))))))))(((((.((....)).)))))..))))))))-- ( -46.30) >consensus GGAAAACCAAAAAGAGG_GGACACUUAACAGGCUUCGUUUGUUGCUAUUUGGCCAGGCCAAACAAACAGGAGUUGGGG____________UCCUCCAAAAGGA_____CUCCCUUCAG__ .............((((.((........(.(((((((((((((......((((...))))))))))).)))))).)...........((((((.......))))))....)))))).... (-14.44 = -16.27 + 1.82)

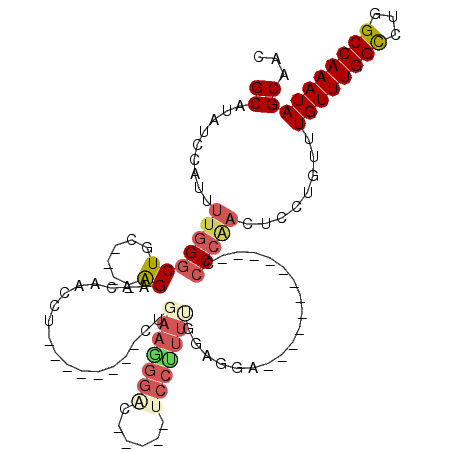
| Location | 12,315,880 – 12,315,971 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -13.03 |
| Energy contribution | -14.43 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12315880 91 - 20766785 GCAUAUCCAUUUUGGGCUGC----AGAACAACCU--------CUGAAGGGAG-----UCCUUUUCGAGGA------------CCCCAACUCCUGUUUGUUUGGCCUGGCCAAAUAGCAAC ((...........(((...(----(((......)--------)))..(((.(-----(((((...)))))------------))))....)))...((((((((...))))))))))... ( -28.00) >DroSec_CAF1 14648 91 - 1 GCAUAUCCAUUUUGGGCUGC----AGAACAACCU--------CUGAAGGGGG-----UCCUUUUGGAGGA------------CCCCAACUCCUGUUUGUUUGGCCUGGCCAAAUAGCAAC ((...........(((((.(----(((......)--------))).))((((-----(((((...)))))------------))))....)))...((((((((...))))))))))... ( -32.80) >DroSim_CAF1 12803 91 - 1 GCAUAUCCAUUUUGGGCUGC----AGAACAACCU--------CUGAAGGGGG-----UCCUUUUGGAGGA------------CCCCAACUCCUGUUUGUUUGGCCUGGCCAAAUAGCAAC ((...........(((((.(----(((......)--------))).))((((-----(((((...)))))------------))))....)))...((((((((...))))))))))... ( -32.80) >DroEre_CAF1 14804 89 - 1 GCAUAUCCAUUUUGGGCUGC----AGAACAACCU--------CUGAAGG-AC-----CCCCUUAGGUGGA------------CCCCGACUCCUGU-UGUUUGGCCUGGCCAAAUAGCAAC .....(((((((.(((((.(----(((......)--------)))..))-.)-----))....)))))))------------..........(((-((((((((...))))))))))).. ( -27.70) >DroYak_CAF1 13092 84 - 1 GCAUAUCCAUUUUUGGCUGC----AGAACAACCUCCAACCCUCUGAAGG-AC-----C-------------------------CCCAACUCCUGU-UGUUUGGCCUGGCCAAAUAGCAAC (((...(((....))).)))----.......(((.((......)).)))-..-----.-------------------------.........(((-((((((((...))))))))))).. ( -20.70) >DroAna_CAF1 12396 111 - 1 GCAUAUCCAUUUCGGACUGCUAGGGGAACCCCCU--------UGGAAAGGACCCUCCUCCUUUCGGCGGAGGAAAAGGACACCCCCACCUCCU-UUUGUUUGGUUUAGCCAAAUAGCAAC ((...(((.....)))...(((((((....))))--------)))((((((.(((((.((....)).)))))....((......))...))))-))(((((((.....)))))))))... ( -37.80) >consensus GCAUAUCCAUUUUGGGCUGC____AGAACAACCU________CUGAAGGGAC_____UCCUUUUGGAGGA____________CCCCAACUCCUGUUUGUUUGGCCUGGCCAAAUAGCAAC ((.........(((((((......))..................(((((((......)))))))...................)))))........((((((((...))))))))))... (-13.03 = -14.43 + 1.41)

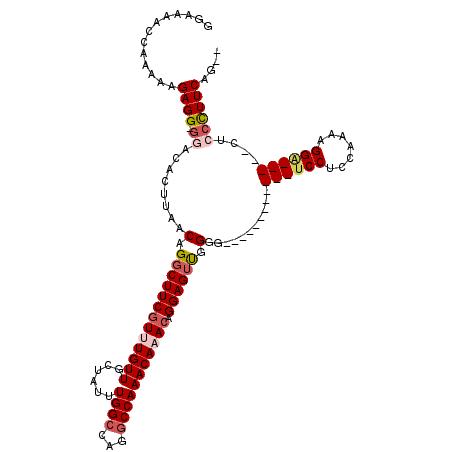
| Location | 12,315,941 – 12,316,038 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.89 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
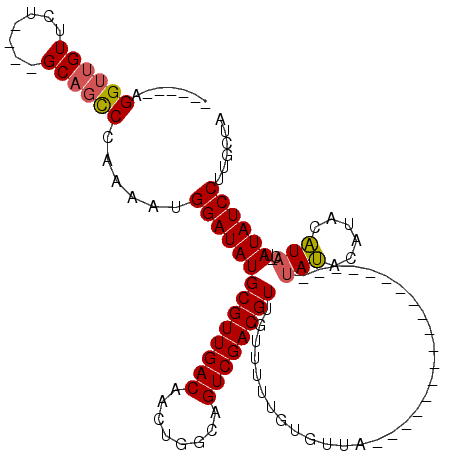
>2R_DroMel_CAF1 12315941 97 + 20766785 ------AGGUUGUUCU----GCAGCCCAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUACUUC-------------AUAUACAUACAUACAUAUAUCCUUGCUA ------.(((((....----.)))))......((((((((((((((........))))))))(((...(((((......-------------..))))).))).....))))))...... ( -26.20) >DroSec_CAF1 14709 92 + 1 ------AGGUUGUUCU----GCAGCCCAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUA------------------UAUAUAUAUAUACUUAUAUCCUUGCUA ------.(((((....----.)))))......((((((((((((((........)))))))).((...(((((..------------------...)))))...))..))))))...... ( -23.80) >DroSim_CAF1 12864 90 + 1 ------AGGUUGUUCU----GCAGCCCAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUA------------------CAUAU--AUAUACAUAUAUCCUUGCUA ------.(((((....----.)))))......((((((((((((((........))))))))(((...(((((..------------------...))--))).))).))))))...... ( -24.30) >DroEre_CAF1 14863 106 + 1 ------AGGUUGUUCU----GCAGCCCAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUACUUCAAUACACACACA--UACAAACUCGUAC--AUAUCCUUGCUG ------.(((((....----.)))))......((((((((((((((........)))))))((((...(((((...........)))))..--.)))).......)--))))))...... ( -28.60) >DroYak_CAF1 13140 116 + 1 GGUUGGAGGUUGUUCU----GCAGCCAAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUACUACCAUACACACACAAAUAUACAUUCAUACGUAUAUCCUUGCUG (((((.(((....)))----.)))))......((((((((((((((........))))))(((((((.(((((...........))))).))))).))........))))))))...... ( -34.90) >DroAna_CAF1 12473 81 + 1 ------AGGGGGUUCCCCUAGCAGUCCGAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUUUC---------------------------------CAUAUCCUUGUUG ------.((((....))))(((((...((.(((((.((((((((((........)))))))).......)).))---------------------------------))).)).))))). ( -23.51) >consensus ______AGGUUGUUCU____GCAGCCCAAAAUGGAUAUGCGUUGACAACUGGCAGUCGACGUUGUUUUUGUGUUA__________________UAUACAUACAUAC_UAUAUCCUUGCUA .......((((((.......))))))......((((((((((((((........))))))))...............................(((......)))...))))))...... (-18.22 = -18.48 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:31 2006