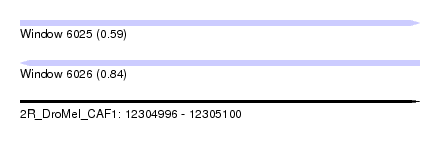
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,304,996 – 12,305,100 |
| Length | 104 |
| Max. P | 0.842350 |

| Location | 12,304,996 – 12,305,100 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
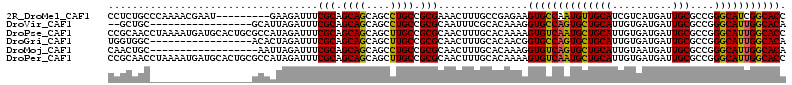
>2R_DroMel_CAF1 12304996 104 + 20766785 CCUCUGCCCAAAACGAAU---------GAAGAUUUCGCAGCAGCAGCCUGCCGCGAAACUUUGCCGAGAAGUGCCAAUGUUGCAUCGUCAUGAUUGCGCCGGGCAUCGGCACC .(((.((...........---------((((.((((((.((((....)))).)))))))))))).)))..(((((.((((((((((.....)).))))....)))).))))). ( -32.50) >DroVir_CAF1 10077 94 + 1 --GCUGC-----------------GCAUUAGAUUUCGCAGCAGCAGCCUGCCGCGCAAUUUCGCACAAAGGUGCCAGUGCUGCAUUGUGAUGAUUGCGCCGGGCAUUGGCACA --(((((-----------------(.(......).))))))....(((((((((((((((...(((((.((((....))))...)))))..))))))))..))))..)))... ( -38.90) >DroPse_CAF1 4181 113 + 1 CCGCAACCUAAAAUGAUGCACUGCGCCAUAGAUUUCGCAGCAGCAGCUUGCCGCGCAACUUUGCACAAAAGUGUCAAUGCUGCAUUGUGAUGAUUGCGCCGGGCAUUGGCACC ..............(((((.((((((.((.....(((((((((((((.....))......((((((....))).)))))))))..)))))..)).))).))))))))...... ( -31.80) >DroGri_CAF1 2305 96 + 1 UGGUGGC-----------------ACACUAGAUUUCGCAGCAGCAGCUUGCCGCGCAACUUUGCACAACGGUGCCAGUGCUGCAUUGUGAUGAUUGCGCCGGGCAUUGGCACA ..(((((-----------------(..((.(....)((....))))..))))))(((....)))......((((((((((((((..........)))....))))))))))). ( -33.10) >DroMoj_CAF1 2007 95 + 1 CAACUGC------------------AAUUAGAUUUCGCAGCAGCAGCCUGCCGCGCAACUUUGCACAAAGGUGUCAGUGCUGCAUUGUAAUGAUUGCGCCGGGCAUUGGCACA ....(((------------------((..((.((.(((.((((....)))).))).))))))))).....((((((((((((((..........)))....))))))))))). ( -30.90) >DroPer_CAF1 1943 113 + 1 CCGCAACCUAAAAUGAUGCACUGCGCCAUAGAUUUCGCAGCAGCAGCUUGCCGCGCAACUUUGCACAAAAGUGUCAAUGCUGCAUUGUGAUGAUUGCGCCGGGCAUUGGCACC ..............(((((.((((((.((.....(((((((((((((.....))......((((((....))).)))))))))..)))))..)).))).))))))))...... ( -31.80) >consensus CCGCUGC_________________GCAAUAGAUUUCGCAGCAGCAGCCUGCCGCGCAACUUUGCACAAAAGUGCCAAUGCUGCAUUGUGAUGAUUGCGCCGGGCAUUGGCACA ...................................(((.((((....)))).)))...............((((((((((((((..........)))....))))))))))). (-26.22 = -25.50 + -0.72)


| Location | 12,304,996 – 12,305,100 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
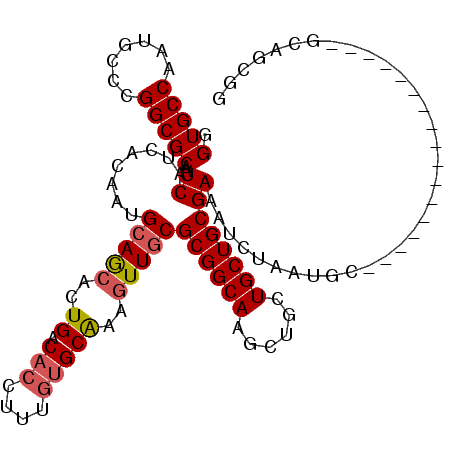
>2R_DroMel_CAF1 12304996 104 - 20766785 GGUGCCGAUGCCCGGCGCAAUCAUGACGAUGCAACAUUGGCACUUCUCGGCAAAGUUUCGCGGCAGGCUGCUGCUGCGAAAUCUUC---------AUUCGUUUUGGGCAGAGG ((((((((((....((...(((.....)))))..)))))))))).(((.((...((((((((((((....))))))))))))..((---------(.......))))).))). ( -39.30) >DroVir_CAF1 10077 94 - 1 UGUGCCAAUGCCCGGCGCAAUCAUCACAAUGCAGCACUGGCACCUUUGUGCGAAAUUGCGCGGCAGGCUGCUGCUGCGAAAUCUAAUGC-----------------GCAGC-- .((((((.(((...(((............))).))).))))))..(((((((......((((((((....))))))))........)))-----------------)))).-- ( -35.14) >DroPse_CAF1 4181 113 - 1 GGUGCCAAUGCCCGGCGCAAUCAUCACAAUGCAGCAUUGACACUUUUGUGCAAAGUUGCGCGGCAAGCUGCUGCUGCGAAAUCUAUGGCGCAGUGCAUCAUUUUAGGUUGCGG ((.((....))))..(((((((........(((((.(((.(((....)))))).)))))((((....)))).((((((..(....)..))))))...........))))))). ( -38.80) >DroGri_CAF1 2305 96 - 1 UGUGCCAAUGCCCGGCGCAAUCAUCACAAUGCAGCACUGGCACCGUUGUGCAAAGUUGCGCGGCAAGCUGCUGCUGCGAAAUCUAGUGU-----------------GCCACCA .............((((((..........(((((((..(((...((((((((....))))))))..)))..)))))))........)))-----------------))).... ( -34.57) >DroMoj_CAF1 2007 95 - 1 UGUGCCAAUGCCCGGCGCAAUCAUUACAAUGCAGCACUGACACCUUUGUGCAAAGUUGCGCGGCAGGCUGCUGCUGCGAAAUCUAAUU------------------GCAGUUG ((((((.......))))))...........(((((.(((......(((((((....))))))))))))))).(((((((.......))------------------))))).. ( -32.60) >DroPer_CAF1 1943 113 - 1 GGUGCCAAUGCCCGGCGCAAUCAUCACAAUGCAGCAUUGACACUUUUGUGCAAAGUUGCGCGGCAAGCUGCUGCUGCGAAAUCUAUGGCGCAGUGCAUCAUUUUAGGUUGCGG ((.((....))))..(((((((........(((((.(((.(((....)))))).)))))((((....)))).((((((..(....)..))))))...........))))))). ( -38.80) >consensus GGUGCCAAUGCCCGGCGCAAUCAUCACAAUGCAGCACUGACACCUUUGUGCAAAGUUGCGCGGCAAGCUGCUGCUGCGAAAUCUAAUGC_________________GCAGCGG .(((((.......)))))..((........(((((..((.(((....)))))..)))))((((((......)))))))).................................. (-25.24 = -25.63 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:25 2006