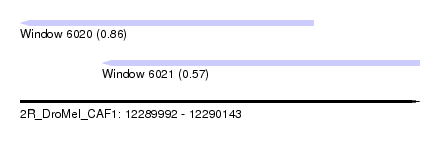
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,289,992 – 12,290,143 |
| Length | 151 |
| Max. P | 0.860151 |

| Location | 12,289,992 – 12,290,103 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.47 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
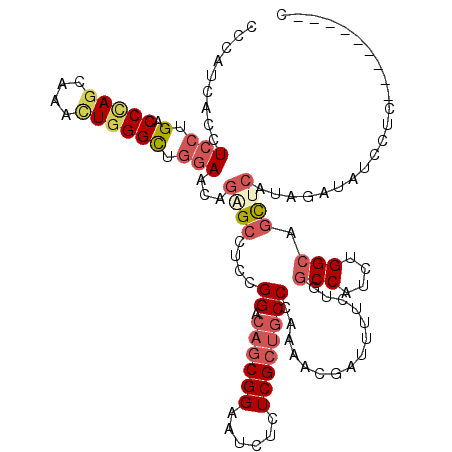
>2R_DroMel_CAF1 12289992 111 - 20766785 CCCAUCACCUCCUGACCCAGCAAACUGGGUUGGAAGAGAGCCUCCGGGCAGCGGAAUCUCUCGCAGCCCAAAACGAUUUUCUGCCCAUCUGGCAGCUCAUAGGUUUCCAC---------C ......((((.(..((((((....))))))..)....((((....((((.((((......)))).))))............((((.....))))))))..))))......---------. ( -40.00) >DroPse_CAF1 715 111 - 1 CCCAGCAUCUGCAGCCCCAGCAGCUUGGGCUGGAACAGGGCCUCCGGACAGCGGAAGCGCUCGCUUCCGAGCUUAAUCCUCUGGCCGUCGGGCAGCUGGUAGACCUCUUC---------G .(((((.((((((((((.((...)).))))))...))))((((.(((.(((.(((...(((((....)))))....))).))).)))..)))).)))))...........---------. ( -46.70) >DroEre_CAF1 2118 111 - 1 CCCAUCACCUCCUGACCCAGCAAACUGGGCUGGAAGAGAGCCUCCGGGCAGCGGAAUCUCUCGCUGCCCAAAACGAUUUUCUGGCCAUCCGGCAGCUCAUAGGUAUCCUC---------C ......((((((.(.(((((....)))))).)))...((((....(((((((((......)))))))))..............(((....))).))))...)))......---------. ( -43.50) >DroYak_CAF1 895 111 - 1 CCCAUCACCUCCUGACCCAGCAAACUGGGCUGGAAGAGAGCCUCCGGACAGCGGAAUCUCUCGCUGCCCAAAACGAUUUUCUGGCCAUCUGGCAGCUCAUAGGUAUCCUC---------C ......((((((.(.(((((....)))))).))).(((((..((((.....))))..)))))((((((.....((......)).......)))))).....)))......---------. ( -36.70) >DroMoj_CAF1 2241 120 - 1 CCGGGCACUUCCAGGCCCAGCAUGUUCGGCUUGAACAGUGCCUCCGGGCAGCGGAAGCGCUCGCUGCCAAUGUUGAUGGUUGUGCCAUCAGGCAGCUCGUAGAACUCAUCGAAAUCCGAC ..(((((((..((((((.((....)).))))))...)))))))..((((.((....))))))((((((.....((((((.....))))))))))))............(((.....))). ( -49.50) >DroAna_CAF1 2134 111 - 1 CCCAUCACCUCCUGACCUAAUAAACUGGGCUGGAACAGUGCCUCCGGACAACGGAAUCUCUCGCUGCCAAGACUGAUCUUCUGGCCAUCUGGCAAUUCGUAGACAUCUUC---------U .....(((.(((.(.((((......))))).)))...)))..((((.....))))...(((((.(((((.((.((..(....)..)))))))))...)).))).......---------. ( -24.30) >consensus CCCAUCACCUCCUGACCCAGCAAACUGGGCUGGAACAGAGCCUCCGGACAGCGGAAUCUCUCGCUGCCCAAAACGAUUUUCUGGCCAUCUGGCAGCUCAUAGAUAUCCUC_________C .........(((.(.(((((....)))))).)))...((((....((.((((((......))))))))...............(((....))).))))...................... (-21.10 = -22.47 + 1.36)

| Location | 12,290,023 – 12,290,143 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.50 |
| Mean single sequence MFE | -46.87 |
| Consensus MFE | -18.50 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
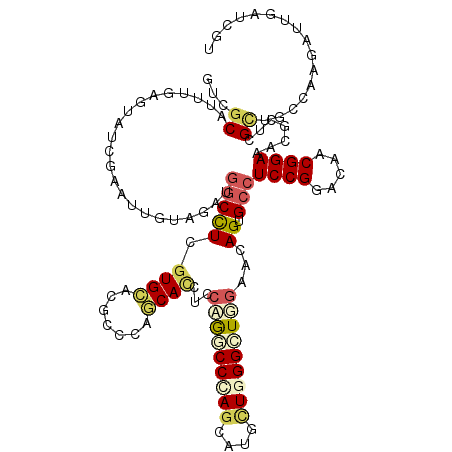
>2R_DroMel_CAF1 12290023 120 - 20766785 GUCGCAGUUGGUUAUCGAGUGAUGGGUGGCCUCGUGGAUGCCCAUCACCUCCUGACCCAGCAAACUGGGUUGGAAGAGAGCCUCCGGGCAGCGGAAUCUCUCGCAGCCCAAAACGAUUUU ((((..(..((((.(((((((((((((..((....))..)))))))).)))(..((((((....))))))..)..)).))))..)((((.((((......)))).))))....))))... ( -54.50) >DroVir_CAF1 2256 120 - 1 AUCGCAUUUGAGUAUUGAGUUAAAGACGGCUUCGUGCAGUCCGGGCACCUCCUGGCCCAGCAUGCUGGGCUUAAAGAGCGCCUCCGGGCAACGGAAACGCUCGUUUCCAAUGUUUAUAGU ...(((((...((((.(((((......))))).)))).(((((((..((((..(((((((....)))))))....))).)..)))))))...((((((....)))))))))))....... ( -44.80) >DroPse_CAF1 746 120 - 1 GUCGCACUGGAGGAUCGACUUGCUGGUGGCCUCGUGCACACCCAGCAUCUGCAGCCCCAGCAGCUUGGGCUGGAACAGGGCCUCCGGACAGCGGAAGCGCUCGCUUCCGAGCUUAAUCCU ...((.(((((((.......((((((((((.....)).)).))))))((((((((((.((...)).))))))...)))).)))))))....(((((((....))))))).))........ ( -51.80) >DroGri_CAF1 1976 120 - 1 AUCACAUUUGAGUAUCGAAUUGUAGAUGUCCUCAUGGACACCGGGCACCUCCAAGCCGAGCAUGCUGGGCUUGAAGAGUGCCUCCGGACAACGGAAACGUUCGCUGCCCACAUUAAUUGU ........((.(((.((((((((...(((((....)))))((((((((((.((((((.((....)).)))))).)).))))..))))))))((....)))))).))).)).......... ( -45.00) >DroMoj_CAF1 2281 120 - 1 GUCGCAUUUCUGUAUCGAAUUGUAGACCGCCUCGUGCAGGCCGGGCACUUCCAGGCCCAGCAUGUUCGGCUUGAACAGUGCCUCCGGGCAGCGGAAGCGCUCGCUGCCAAUGUUGAUGGU ((((((.(((......))).))).))).((((((.(((((..(((((((..((((((.((....)).))))))...))))))))).((((((((......))))))))..)))))).))) ( -46.70) >DroAna_CAF1 2165 119 - 1 GUCGCAGUG-GAGGAGGACUGGUAAGUGGCCUCGUGGAUGCCCAUCACCUCCUGACCUAAUAAACUGGGCUGGAACAGUGCCUCCGGACAACGGAAUCUCUCGCUGCCAAGACUGAUCUU (((((((((-..(((((((((....(.(((.((...)).))))......(((.(.((((......))))).))).)))).))((((.....)))).)))..))))))...)))....... ( -38.40) >consensus GUCGCAUUUGAGUAUCGAAUUGUAGAUGGCCUCGUGCACGCCCAGCACCUCCAGGCCCAGCAUGCUGGGCUGGAACAGUGCCUCCGGACAACGGAAACGCUCGCUGCCAAGAUUGAUCGU ...((......................(((((.((((.......))))...(((((((((....)))))))))...)).)))((((.....)))).......))................ (-18.50 = -19.20 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:20 2006