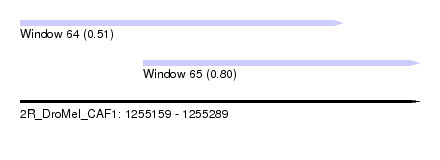
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,255,159 – 1,255,289 |
| Length | 130 |
| Max. P | 0.795467 |

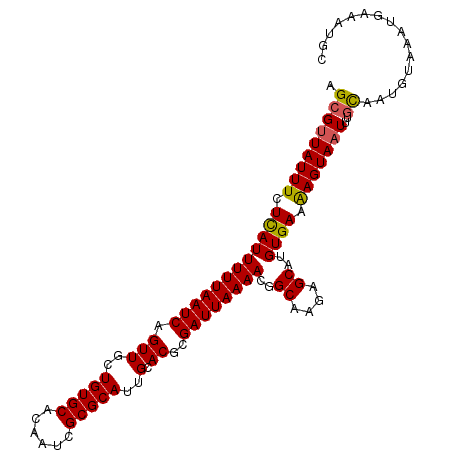
| Location | 1,255,159 – 1,255,264 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
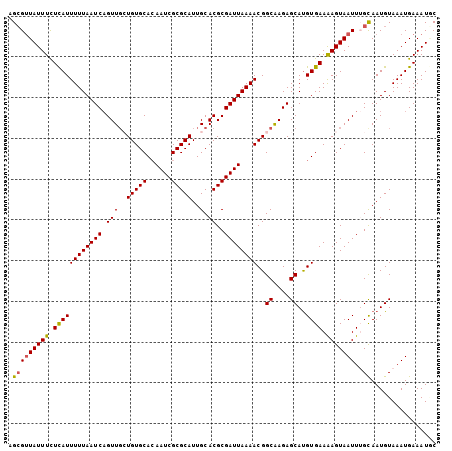
>2R_DroMel_CAF1 1255159 105 + 20766785 AGCGUUAUUUCUCAUUUUUAAUCAGUUUCUGUGCACAAUCGCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGC .(((((.....((((((..........(((.(((..(((((((.......)))))))......))))))(((((..((......))..).))))))))))))))) ( -20.60) >DroSec_CAF1 6890 105 + 1 CGCGUUAUUUCUCAUUUUUAAUCAGUUGCUGUGCACAAUCGCGCAUUGCACGCGAUUAAAACGGCAAGAGCACGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGC .(((((.....((((((.((.....(((((((....(((((((.......)))))))...)))))))..(((.((........)).)))..)).))))))))))) ( -25.40) >DroSim_CAF1 12716 105 + 1 AGCGUUAUUUCUCAUUUUUAAUCAGUUGCUGUGCACAAUCGCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUUC .(((((((((.((((......((..(((((((....(((((((.......)))))))...)))))))))....)))).)))))))..))................ ( -25.60) >DroEre_CAF1 11119 105 + 1 AGCGGUAUUUCUUAUUUUUAAUCAGUUGGUGUGCACAAUCGCGCAUUGCACGCGAUUAAAACGGCGAGAGCAUGUGAAAAGUAAUUAGCAAUGUAAAUGAAAUAC ....(((((((.....(((((((.((((((((((......)))))))).))..)))))))(((((....)).))).......................))))))) ( -24.00) >DroYak_CAF1 10403 105 + 1 AGAGUUAUUUUUUAUUUUUAAUCCGUUGGUGUGCAGAAUCGCGCAUUGCACGCGAUUAAAACGGCGAGAGCAUGUGAAGAGUAAUUAGUAAUGUAAAUGAAAUUC ..((((((((((((((((((((((((((((((((......)))))))).))).))))))))..((....))..)))))))))))))................... ( -29.50) >consensus AGCGUUAUUUCUCAUUUUUAAUCAGUUGCUGUGCACAAUCGCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGC .(((((((((.((((((((((((.(((..(((((......)))))..).))..))))))))..((....))..)))).)))))))..))................ (-19.78 = -19.62 + -0.16)

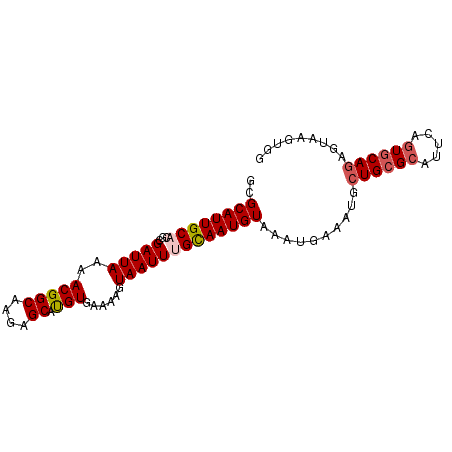
| Location | 1,255,199 – 1,255,289 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1255199 90 + 20766785 GCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGCUGCGCAUUCAGUGCACAGUAAGUGG ..((((((((...(((((..(((((....)).)))......)))))))))))))........(((((.((((...)))).)))))..... ( -22.20) >DroSec_CAF1 6930 90 + 1 GCGCAUUGCACGCGAUUAAAACGGCAAGAGCACGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGCUGCGCAUUCAGUGCAGAGUAAGUGG ..((((((((...(((((..(((((....)).)))......)))))))))))))..........((((((.....))))))......... ( -23.70) >DroSim_CAF1 12756 90 + 1 GCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUUCUGCGCAUUCAAUGCAGAGUAAGUGG ..((((((((...(((((..(((((....)).)))......))))))))))))).......((((((((.......))))))))...... ( -20.90) >DroEre_CAF1 11159 90 + 1 GCGCAUUGCACGCGAUUAAAACGGCGAGAGCAUGUGAAAAGUAAUUAGCAAUGUAAAUGAAAUACUGCGCAUUCAGUGCAGAGUAAGUGG ..(((((((....(((((..(((((....)).)))......))))).)))))))..........((((((.....))))))......... ( -22.60) >DroYak_CAF1 10443 89 + 1 GCGCAUUGCACGCGAUUAAAACGGCGAGAGCAUGUGAAGAGUAAUUAGUAAUGUAAAUGAAAUUCUGCGCAUACAGU-CAGAGUAAGUGG ..(((((((.....(((...(((((....)).)))....))).....))))))).......((((((.((.....))-))))))...... ( -17.70) >consensus GCGCAUUGCACGCGAUUAAAACGGCAAGAGCAUGUGAAAAGUAAUUUGCAAUGUAAAUGAAAUGCUGCGCAUUCAGUGCAGAGUAAGUGG ..((((((((...(((((..(((((....)).)))......)))))))))))))..........((((((.....))))))......... (-18.58 = -19.26 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:10 2006