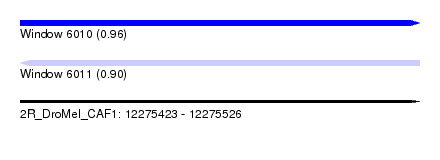
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,275,423 – 12,275,526 |
| Length | 103 |
| Max. P | 0.962843 |

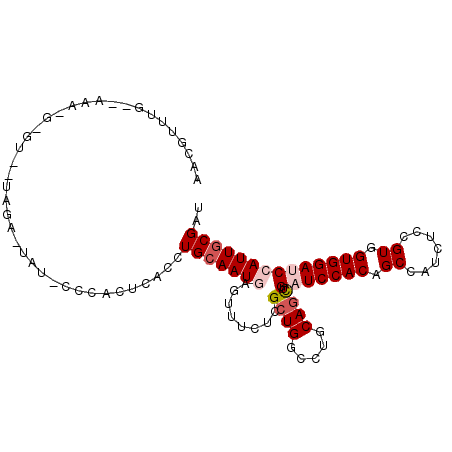
| Location | 12,275,423 – 12,275,526 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.28 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12275423 103 + 20766785 AACGUUUGGUAAAGGGGUCUUAGA-UAUUCCCACUCACCUGCAAUAAGUUUCUCGCUGGCCUGCAGCUGAUCCACAGCCAUCUCCGUGGUGGAUCCAUUACGAU ......((((...((((.......-...))))..(((.(((((...(((.....)))....))))).)))......))))....(((((((....))))))).. ( -26.40) >DroVir_CAF1 167461 90 + 1 UGGAUUGC-------------CUC-UAGGCGAGCUUACCUGCAAUCAGCUUCUCGCUGGCCUGCAAUUGAUCCACGGCCAUCUCCGUGGUGGAACCAUUGCGAU .((.((((-------------(..-..)))))((......))..(((((.....)))))))((((((.(.(((((.((.......)).))))).).)))))).. ( -29.60) >DroGri_CAF1 171979 84 + 1 AGCGUUUG-------------AUU-------UGCUUACCUGCAAUGAGCUUCUCGCUGGCCUGCAGCUGAUCCACGGCCAUUUCGGUGGUGGAACCAUUGCGAU ((((....-------------...-------))))....(((((((........((((.....))))...(((((.(((.....))).)))))..))))))).. ( -28.10) >DroSim_CAF1 132928 91 + 1 ----------AAGGGAGUUAGAU--UAU-UCCACUCACCUGCAAUGAGUUUCUCGCUGGCCUGCAGCUGAUCCACAGCCAUCUCCGUUGUGGAUCCAUUGCGAU ----------.(((((((..((.--...-)).)))).)))((((((........((((.....)))).((((((((((.......))))))))))))))))... ( -34.20) >DroEre_CAF1 145471 104 + 1 CACGUUUGGUAAAAGAGCGUUAGAUGAUCCACACUCACCUGCAAUGAGUUUCUCGCUGGCCUGCAGCUGAUCCACAGCCAUCUCCGUUGUGGAUCCAUUGCGAU .((((((.......)))))).((.(((.......))).))((((((........((((.....)))).((((((((((.......))))))))))))))))... ( -31.60) >DroYak_CAF1 141053 104 + 1 CACGUUUGUUAAAAGCUUCAUAGAUUAUCCCCACUUACCUGCAAUGAGUUUCUCGCUGGCCUGCAGCUGAUCCACAGCCAUUUCCGUUGUGGAUCCAUUGCGAU ...((((.....)))).......................(((((((........((((.....)))).((((((((((.......))))))))))))))))).. ( -28.90) >consensus AACGUUUG__AAA_G_GU__UAGA_UAU_CCCACUCACCUGCAAUGAGUUUCUCGCUGGCCUGCAGCUGAUCCACAGCCAUCUCCGUGGUGGAUCCAUUGCGAU .......................................(((((((........((((.....)))).(((((((.((.......)).)))))))))))))).. (-19.37 = -20.23 + 0.86)

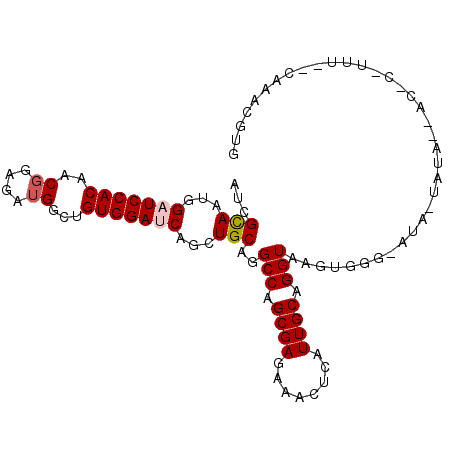
| Location | 12,275,423 – 12,275,526 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 74.28 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12275423 103 - 20766785 AUCGUAAUGGAUCCACCACGGAGAUGGCUGUGGAUCAGCUGCAGGCCAGCGAGAAACUUAUUGCAGGUGAGUGGGAAUA-UCUAAGACCCCUUUACCAAACGUU ...(((((((((((((..((....))...))))))).((((.....))))........)))))).((((((.(((....-.......))).))))))....... ( -30.00) >DroVir_CAF1 167461 90 - 1 AUCGCAAUGGUUCCACCACGGAGAUGGCCGUGGAUCAAUUGCAGGCCAGCGAGAAGCUGAUUGCAGGUAAGCUCGCCUA-GAG-------------GCAAUCCA ...((..........((((((......)))))).....((((((..((((.....)))).))))))....))..(((..-..)-------------))...... ( -30.00) >DroGri_CAF1 171979 84 - 1 AUCGCAAUGGUUCCACCACCGAAAUGGCCGUGGAUCAGCUGCAGGCCAGCGAGAAGCUCAUUGCAGGUAAGCA-------AAU-------------CAAACGCU ...((..((((.((((..((.....))..)))))))).((((((...(((.....)))..))))))....)).-------...-------------........ ( -23.30) >DroSim_CAF1 132928 91 - 1 AUCGCAAUGGAUCCACAACGGAGAUGGCUGUGGAUCAGCUGCAGGCCAGCGAGAAACUCAUUGCAGGUGAGUGGA-AUA--AUCUAACUCCCUU---------- ...((((((((((((((.((....))..)))))))).((((.....))))........))))))(((.(((((((-...--.))).))))))).---------- ( -33.50) >DroEre_CAF1 145471 104 - 1 AUCGCAAUGGAUCCACAACGGAGAUGGCUGUGGAUCAGCUGCAGGCCAGCGAGAAACUCAUUGCAGGUGAGUGUGGAUCAUCUAACGCUCUUUUACCAAACGUG ...((((((((((((((.((....))..)))))))).((((.....))))........)))))).(((((((((((.....)).))))))....)))....... ( -35.30) >DroYak_CAF1 141053 104 - 1 AUCGCAAUGGAUCCACAACGGAAAUGGCUGUGGAUCAGCUGCAGGCCAGCGAGAAACUCAUUGCAGGUAAGUGGGGAUAAUCUAUGAAGCUUUUAACAAACGUG ...((((((((((((((.((....))..)))))))).((((.....))))........))))))((((..(((((.....)))))...))))............ ( -29.40) >consensus AUCGCAAUGGAUCCACAACGGAGAUGGCUGUGGAUCAGCUGCAGGCCAGCGAGAAACUCAUUGCAGGUAAGUGGG_AUA_UAUA__AC_C_UUU__CAAACGUG ...(((...(((((((..((....))...)))))))...)))..(((.((((........)))).))).................................... (-19.56 = -19.92 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:10 2006