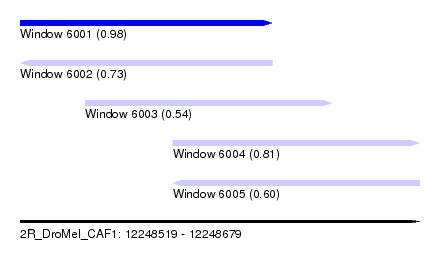
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,248,519 – 12,248,679 |
| Length | 160 |
| Max. P | 0.977762 |

| Location | 12,248,519 – 12,248,620 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -22.41 |
| Energy contribution | -23.64 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
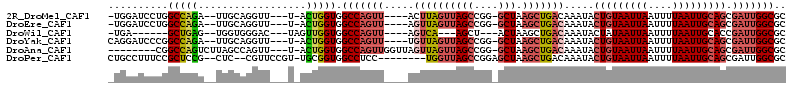
>2R_DroMel_CAF1 12248519 101 + 20766785 -UGGAUCCUGGCCAGA--UUGCAGGUU---U-ACUGGUGGCCAGUU----ACUUAGUUAGCCGG-GCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC -.........(((((.--((((((((.---.-(((((...))))).----)))).((((((...-.....))))))......((((((((....))))))))))))))))).. ( -30.60) >DroEre_CAF1 117596 101 + 1 -UGGAUCCUGGCCAGA--UUGCAGGUU---U-ACUGGUGGCCAGUU----AGUUAGUUAGCCGG-GCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC -..(((((((((((..--...(((...---.-.))).)))))))..----.((((((((((...-))).))))))).....(((((((((....))))))))).))))..... ( -31.40) >DroWil_CAF1 172747 91 + 1 -UGA------GCUGAG--UGGUGGGAC---UAGUUGGUGGCCAGUU----AGUCA---AGCU---ACUAAGCUGACAAAUACUAUAAUUAAUUUUAAUUGCACCGAUUGGCGC -..(------(((.((--((((..(((---((..(((...)))..)----)))).---.)))---))).))))..........................((.((....)).)) ( -26.80) >DroYak_CAF1 112716 102 + 1 CAGGAUCCCGGCCAGA--UUGCAGGUU---U-ACUGGUGGCCAGUU----UGUUAGUUAGCCGG-GCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC (((.(..((.((....--..)).))..---)-.)))...(((((((----(((((((((((...-))).))))))))))).(((((((((....)))))))))....)))).. ( -33.00) >DroAna_CAF1 99823 100 + 1 --------CGGCCAGUCUUAGCCAGUU---U-ACUGGUGGCCAGUUGGUUAGUUAGUUAGCCGG-GCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC --------..(((((((.((((((...---.-(((((...)))))))))))((((((((((...-))).))))))).....(((((((((....))))))))).))))))).. ( -37.50) >DroPer_CAF1 116503 100 + 1 CUGCCUUUCCGCUCCG--CUC--CGUUCCGU-UGCGGUGGCCUCC--------UGGUUAGCCGGAGCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC ..(((..((.(((..(--(((--(((.....-.))(((((((...--------.))))).)))))))..))).........(((((((((....))))))))).))..))).. ( -36.00) >consensus _UGGAUCCCGGCCAGA__UUGCAGGUU___U_ACUGGUGGCCAGUU____AGUUAGUUAGCCGG_GCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGC ..........(((((..................))))).(((((((.....((((((((((....))).))))))).....(((((((((....))))))))).))))))).. (-22.41 = -23.64 + 1.22)
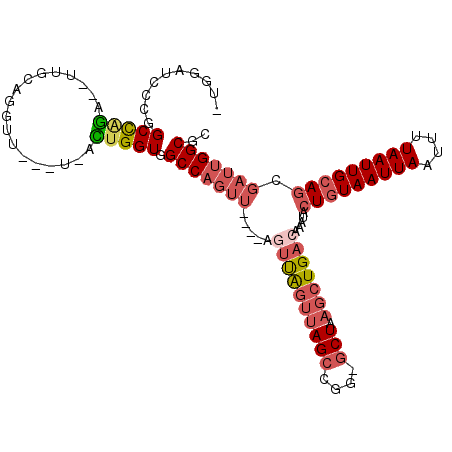
| Location | 12,248,519 – 12,248,620 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.93 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12248519 101 - 20766785 GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGC-CCGGCUAACUAAGU----AACUGGCCACCAGU-A---AACCUGCAA--UCUGGCCAGGAUCCA- ..(.(((..((((.(((((....))))).))))..))).).((((((.-........))))))----..(((((((...((-(---....)))..--..)))))))......- ( -24.60) >DroEre_CAF1 117596 101 - 1 GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGC-CCGGCUAACUAACU----AACUGGCCACCAGU-A---AACCUGCAA--UCUGGCCAGGAUCCA- ((..(((..((((.(((((....))))).))))..)))...))(((((-...)))))......----..(((((((...((-(---....)))..--..)))))))......- ( -24.40) >DroWil_CAF1 172747 91 - 1 GCGCCAAUCGGUGCAAUUAAAAUUAAUUAUAGUAUUUGUCAGCUUAGU---AGCU---UGACU----AACUGGCCACCAACUA---GUCCCACCA--CUCAGC------UCA- (((((....)))))..........................((((.(((---.(..---.((((----(..(((...)))..))---)))...).)--)).)))------)..- ( -20.40) >DroYak_CAF1 112716 102 - 1 GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGC-CCGGCUAACUAACA----AACUGGCCACCAGU-A---AACCUGCAA--UCUGGCCGGGAUCCUG ((..(((..((((.(((((....))))).))))..)))...))....(-(((((((.....((----.(((((...)))))-.---....))...--..))))))))...... ( -26.80) >DroAna_CAF1 99823 100 - 1 GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGC-CCGGCUAACUAACUAACCAACUGGCCACCAGU-A---AACUGGCUAAGACUGGCCG-------- .........((((.(((((....))))).))))....(((((((((((-(.((.((......)).)).(((((...)))))-.---....))))))).)))))..-------- ( -28.00) >DroPer_CAF1 116503 100 - 1 GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGCUCCGGCUAACCA--------GGAGGCCACCGCA-ACGGAACG--GAG--CGGAGCGGAAAGGCAG ..(((....((((.(((((....))))).)))).....((.((((.(((((((((.....--------...)))).(((..-.)))...)--)))--).)))).))..))).. ( -35.10) >consensus GCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUCAGCUUAGC_CCGGCUAACUAACU____AACUGGCCACCAGU_A___AACCUGCAA__UCUGGCCGGGAUCCA_ ((..(((..((((.(((((....))))).))))..)))...))(((((....)))))..............(((((.......................)))))......... (-11.71 = -12.93 + 1.22)
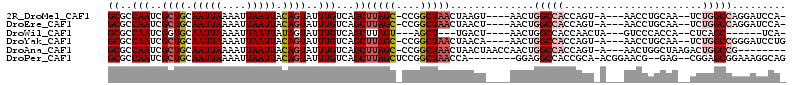
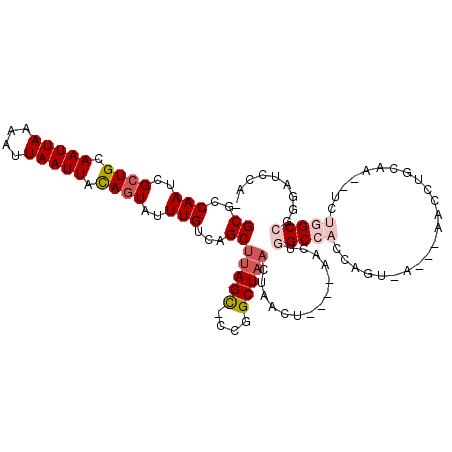
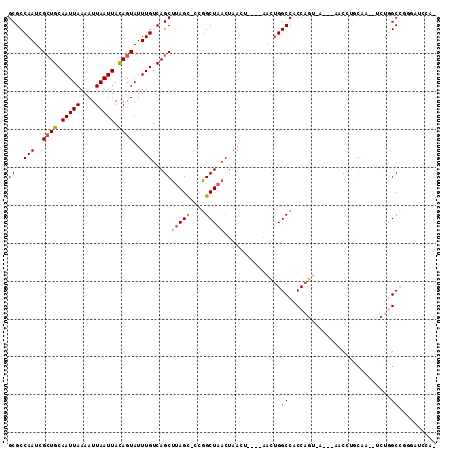
| Location | 12,248,545 – 12,248,644 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12248545 99 + 20766785 CUGGUGGCCAGUUACUUAGUUAGCCGGGCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUACAGACU-------------- ..(((((((((((.....((((((........)))))).....(((((((((....))))))))).)))))))...)))).--...((((......)))).-------------- ( -28.30) >DroVir_CAF1 138674 88 + 1 UUGGUGGCCAGUUAG--------CCA--CAAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUU--UCCGUCUCAUUAAG---UUC------------ ..((..(((((((((--------(..--....)))))).....(((((((((....)))))))))....))))..))....--..............---...------------ ( -24.20) >DroGri_CAF1 143222 83 + 1 UUGGUGGCCAGUUA------------------GCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUU--UCCGUCUCAUUAAAGGUAUU------------ ..((..(((((((.------------------((((.(((.((..((.....))..)).))))))))))))))..))((((--(...........)))))...------------ ( -18.90) >DroEre_CAF1 117622 113 + 1 CUGGUGGCCAGUUAGUUAGUUAGCCGGGCUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUU--UCCGUCUCAUUAAAGACUGCAGGCCCCCAACU .(((.((((.(((((((.((((((........)))))).....(((((((((....))))))))).)))))))....((..--...((((......)))).)).)))).)))... ( -36.10) >DroWil_CAF1 172768 94 + 1 UUGGUGGCCAGUUAGUCA---AGCU--ACUAAGCUGACAAAUACUAUAAUUAAUUUUAAUUGCACCGAUUGGCGCUCUCUU--UCCGUCUCAUUAAAGUAU-------------- (((((((((((((.((((---.(((--....)))))))........((((((....))))))....)))))))((......--...))..)))))).....-------------- ( -17.60) >DroMoj_CAF1 148977 94 + 1 GUGGUGGCCAGUUAG-------GCCA--GAAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUUUUUCCGUCUCAUUAAAGUUUGC------------ ((((..(((((((.(-------((..--....)))........(((((((((....))))))))).)))))))..))))........................------------ ( -25.00) >consensus UUGGUGGCCAGUUAG_______GCC___CUAAGCUGACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCCCGCUU__UCCGUCUCAUUAAAGACU_C____________ ..((((.((((((.........((........)).........(((((((((....))))))))).))))))))))....................................... (-15.02 = -15.60 + 0.58)
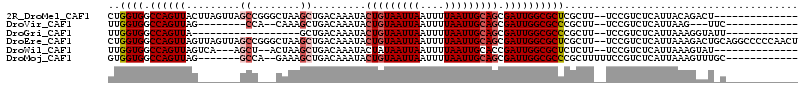
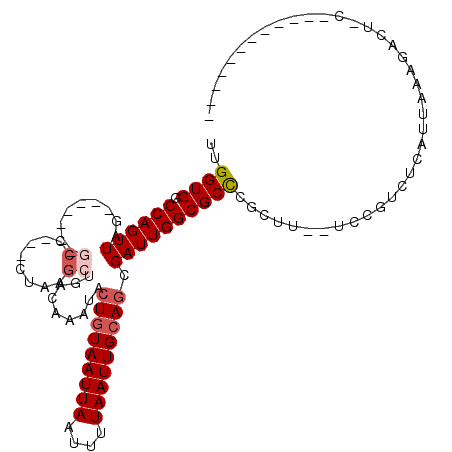
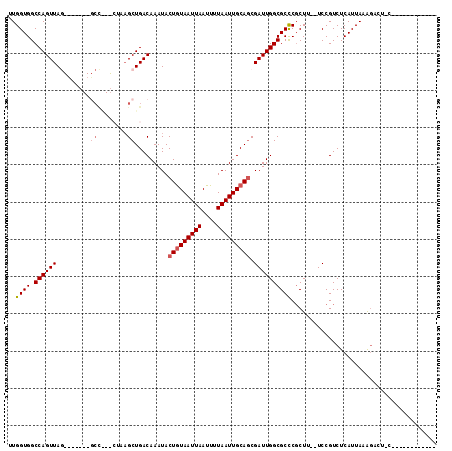
| Location | 12,248,580 – 12,248,679 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12248580 99 + 20766785 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUACAGACU--------------GCACUGCUCCCUUUUUGUU--UUUUUUUUUUUACUA---C ((((((..(((((((((....))))))))).....((.((..((.....((((......)))).--------------))...)).))...))))))--...............---. ( -17.60) >DroSec_CAF1 112748 82 + 1 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUACAGACU--------------GCACUGCUCCCUUUUUUU---------------------- ........(((((((((....))))))))).....((.((..((.....((((......)))).--------------))...)).))........---------------------- ( -14.90) >DroSim_CAF1 106326 82 + 1 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUACAGACU--------------GCACUGCUCCCUUUUUUU---------------------- ........(((((((((....))))))))).....((.((..((.....((((......)))).--------------))...)).))........---------------------- ( -14.90) >DroEre_CAF1 117657 99 + 1 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUAAAGACUGCAGGCCCCCAACUGCACUGCUCCCUUUUAUUU--U-----------------C (((.....(((((((((....))))))))).((..(((....))).)).))).....((((...((((((........)).))))...)))).....--.-----------------. ( -21.50) >DroYak_CAF1 112778 95 + 1 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUAAAGACU--------------GCACUGCUCCCUUUUUUUU--UAUGCCUUCUU-------A (((.....(((((((((....))))))))).((..(((....))).)).)))......((((..--------------(((................--..)))..))))-------. ( -15.97) >DroAna_CAF1 99883 111 + 1 GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUACAGACUGUAGGCU-------GCCGUGCUCCCUUUUUGCUCCUUUUUUUUUUUAGUCAGUC ........(((((((((....))))))))).(((((((((..((((.(.((((......)))).).)))).-------)).(.((.........)).).............))))))) ( -24.80) >consensus GACAAAUACUGUAAUUAAUUUUAAUUGCAGCGAUUGGCGCUCGCUUUCCGUCUCAUUACAGACU______________GCACUGCUCCCUUUUUUUU__U_________________C (((.....(((((((((....))))))))).((..(((....))).)).))).................................................................. (-13.40 = -13.40 + 0.00)

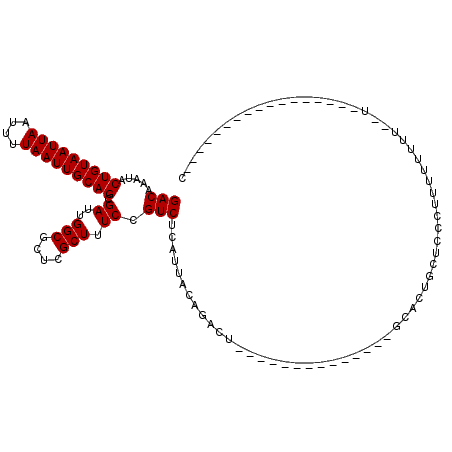
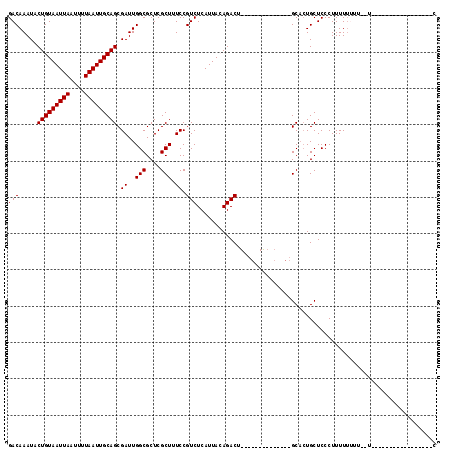
| Location | 12,248,580 – 12,248,679 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12248580 99 - 20766785 G---UAGUAAAAAAAAAAA--AACAAAAAGGGAGCAGUGC--------------AGUCUGUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC .---...............--.(((((...((.((..(((--------------..(((((......)))))..))).)).))....((((.(((((....))))).)))).))))). ( -20.40) >DroSec_CAF1 112748 82 - 1 ----------------------AAAAAAAGGGAGCAGUGC--------------AGUCUGUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC ----------------------........((.((..(((--------------..(((((......)))))..))).)).))....((((.(((((....))))).))))....... ( -18.40) >DroSim_CAF1 106326 82 - 1 ----------------------AAAAAAAGGGAGCAGUGC--------------AGUCUGUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC ----------------------........((.((..(((--------------..(((((......)))))..))).)).))....((((.(((((....))))).))))....... ( -18.40) >DroEre_CAF1 117657 99 - 1 G-----------------A--AAAUAAAAGGGAGCAGUGCAGUUGGGGGCCUGCAGUCUUUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC .-----------------.--............((((((..(((((.(.(.(((.(((((....))))).....))).)).))))))))))).......................... ( -23.10) >DroYak_CAF1 112778 95 - 1 U-------AAGAAGGCAUA--AAAAAAAAGGGAGCAGUGC--------------AGUCUUUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC .-------.....((((..--.........((.((.((.(--------------.(((((....))))).)...))..)).))....((((.(((((....))))).))))...)))) ( -18.60) >DroAna_CAF1 99883 111 - 1 GACUGACUAAAAAAAAAAAGGAGCAAAAAGGGAGCACGGC-------AGCCUACAGUCUGUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC .((((..............(.(((.....(....)..(((-------.((......(((((......)))))......)))))....))).)(((((....))))).))))....... ( -20.10) >consensus G_________________A__AAAAAAAAGGGAGCAGUGC______________AGUCUGUAAUGAGACGGAAAGCGAGCGCCAAUCGCUGCAAUUAAAAUUAAUUACAGUAUUUGUC .................................((((((................((((......))))((...(....).))...)))))).......................... (-13.40 = -13.73 + 0.33)

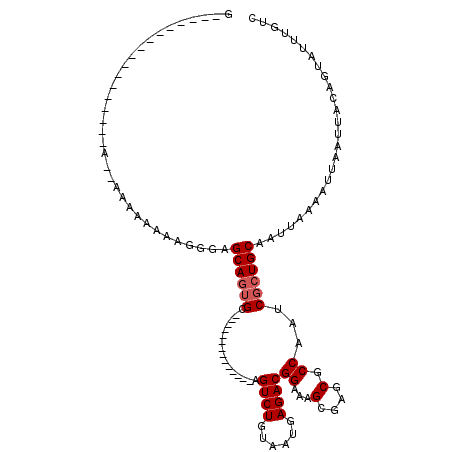
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:04 2006