
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,238,013 – 12,238,177 |
| Length | 164 |
| Max. P | 0.903932 |

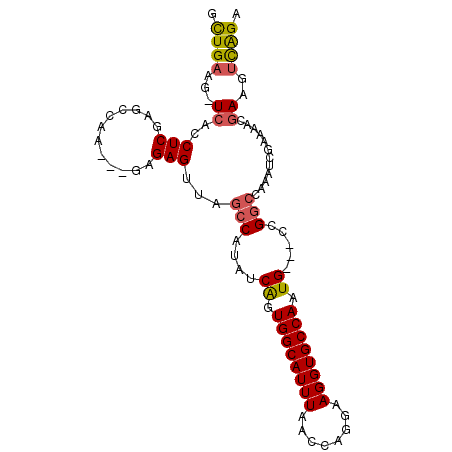
| Location | 12,238,013 – 12,238,108 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -14.78 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
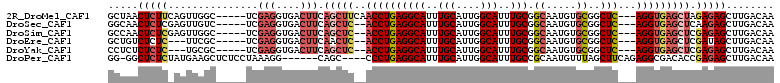
>2R_DroMel_CAF1 12238013 95 - 20766785 GCUGAAG-UCACCUCGAGCCAACUGAAGAGUUAGCCAUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUG---CCGGCCAAAUCGAAAACGAAGUCAGU (((((..-((...(((((((.(((((.............)))))((((((..(((.....)))...))))---)))))....))))....))..))))) ( -28.62) >DroSec_CAF1 98955 95 - 1 GCUGAAG-UCACCUCGAGACAACUCGAGAGUUGCCCAUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUG---UCGGCCAAUUCGAAAACUAAGUCAGA .((((..-.....(((((....)))))((((((((.((((...((((((((........)))))))))))---).)).))))))..........)))). ( -27.70) >DroSim_CAF1 95944 95 - 1 GCUGAAG-UCACCUCGAGCCAACUCGAGAGUUGGCCAUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUG---UCGGCCAAAUCGAAAACGAAGUCAGA .((((..-((..((((((....))))))..((((((....((.((((((((........)))))))).))---..)))))).........))..)))). ( -35.60) >DroEre_CAF1 106897 92 - 1 GUUGAAG-UCACCUCGAGCGAA---GAGAGACAGCCAUAUCGGUGGCAUUUAACCAGGAAGGUGCCAGUG---CUGGCCAAAUCGAAAACGAAGUCAGA .((((..-((..(((.......---))).....((((...((.((((((((........)))))))).))---.))))............))..)))). ( -23.80) >DroYak_CAF1 102201 92 - 1 GCUGAAG-UCACCUCGAGCGCA---GAGAGAGAGGCAUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUG---CCGGCCAAAUCGAAAACGAAGUCAGA .((((..-((..(((.......---))).((..(((....((.((((((((........)))))))).))---...)))...))......))..)))). ( -25.80) >DroAna_CAF1 90866 80 - 1 GCUGUGGCUCAC-------------------GGGCCGUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUGAGGUUGGCAUUUUUAGAUACGAAUUUGGG .....((((...-------------------.))))(((((..(((.......))).((((((((((((...)))))))))))).)))))......... ( -26.30) >consensus GCUGAAG_UCACCUCGAGCCAA___GAGAGUUAGCCAUAUCAGUGGCAUUUAACCAGGAAGGUGCCAAUG___CCGGCCAAAUCGAAAACGAAGUCAGA .((((...((..(((............)))...(((....((.((((((((........)))))))).)).....)))............))..)))). (-14.78 = -15.02 + 0.24)

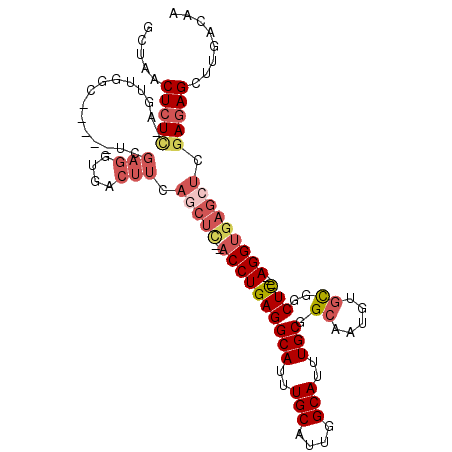
| Location | 12,238,074 – 12,238,177 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.83 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
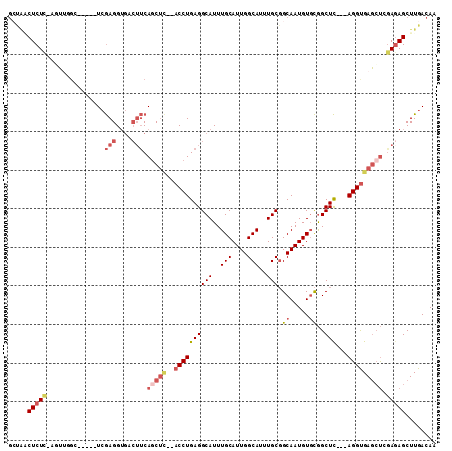
>2R_DroMel_CAF1 12238074 103 + 20766785 GCUAACUCUUCAGUUGGC-----UCGAGGUGACUUCAGCUUCAACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC---AGGUGAGCUAGAGAGCUUGACAA (((((((....)))))))-----(((((.(..((..(((((..((((((((((..(((....)))..))).((.....))..)))---)))))))))..))).)))))... ( -39.90) >DroSec_CAF1 99016 101 + 1 GGCAACUCUCGAGUUGUC-----UCGAGGUGACUUCAGCUC--ACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC---AGGUGAGCUCAAGAGCUUGACAA (((.((.((((((....)-----)))))))..(((.(((((--((((((((((..(((....)))..))).((.....))..)))---))))))))).))).)))...... ( -47.20) >DroSim_CAF1 96005 101 + 1 GCCAACUCUCGAGUUGGC-----UCGAGGUGACUUCAGCUC--ACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC---AGGUGAGCUCGAGAGCUUGACAA ..((((((((((((((((-----(.(((....))).))))(--((((((((((..(((....)))..))).((.....))..)))---))))))))))))))).))).... ( -50.30) >DroEre_CAF1 106958 98 + 1 GCUGUCUCUC---UUCGC-----UCGAGGUGACUUCAACUC--ACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC---AGGUGAGCUCGAUAGCUUGACAA ((((((((.(---(((..-----..)))).))......(((--((((((((((..(((....)))..))).((.....))..)))---)))))))...))))))....... ( -40.50) >DroYak_CAF1 102262 98 + 1 CCUCUCUCUC---UGCGC-----UCGAGGUGACUUCAGCUC--ACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC---AGGUGAGCUCGAGAGCUUGACAA ......((..---.((.(-----(((((....)....((((--((((((((((..(((....)))..))).((.....))..)))---))))))))))))).))..))... ( -43.10) >DroPer_CAF1 103082 100 + 1 GG-GGCUCUCUAUGAAGCUCUCCUAAAGG------CAGC----CCCUGAGGCAUUUGCAUUGGCAUUUGCCGCAAUGUUUAGCUUCAGAGGCGACACCGAGAGCUUGACAA .(-(((((((..((..(((........))------).((----(.(((((((....((((((((....)))..)))))...))))))).)))..))..))))))))..... ( -39.40) >consensus GCUAACUCUC_AGUUGGC_____UCGAGGUGACUUCAGCUC__ACCUGAGGCAUUUGCAUUGGCAUUUGCGGCAAUGUGCGGCUC___AGGUGAGCUCGAGAGCUUGACAA .......................(((((.(..(((.(((((..((((((((((..(((....)))..))).((.....))..)))...))))))))).)))).)))))... (-19.14 = -20.83 + 1.70)
| Location | 12,238,074 – 12,238,177 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -37.25 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.93 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
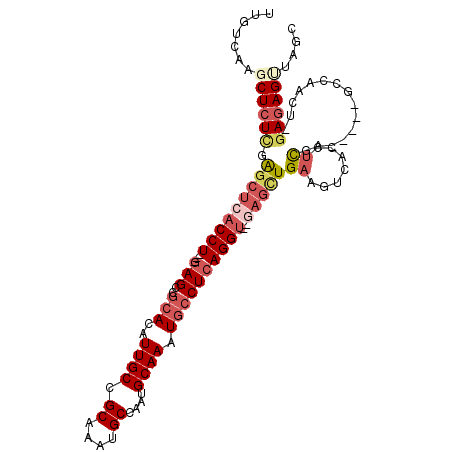
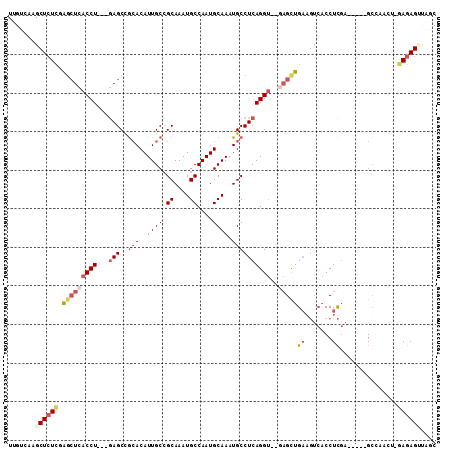
>2R_DroMel_CAF1 12238074 103 - 20766785 UUGUCAAGCUCUCUAGCUCACCU---GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGUUGAAGCUGAAGUCACCUCGA-----GCCAACUGAAGAGUUAGC ......((((((.(((.((((((---(((..(((..((((.((....))....)))).))))))))).))).(((..((....))..)-----))...))).))))))... ( -27.30) >DroSec_CAF1 99016 101 - 1 UUGUCAAGCUCUUGAGCUCACCU---GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGU--GAGCUGAAGUCACCUCGA-----GACAACUCGAGAGUUGCC .......((.(((.(((((((((---(((..(((..((((.((....))....)))).))))))))))--))))).)))..(((((((-----(....)))))).)).)). ( -41.70) >DroSim_CAF1 96005 101 - 1 UUGUCAAGCUCUCGAGCUCACCU---GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGU--GAGCUGAAGUCACCUCGA-----GCCAACUCGAGAGUUGGC ..(((((.(((((.(((((((((---(((..(((..((((.((....))....)))).))))))))))--)))))...........((-----(....))))))))))))) ( -42.50) >DroEre_CAF1 106958 98 - 1 UUGUCAAGCUAUCGAGCUCACCU---GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGU--GAGUUGAAGUCACCUCGA-----GCGAA---GAGAGACAGC (((((..(((.((.(((((((((---(((..(((..((((.((....))....)))).))))))))))--))))))))))...(((..-----.....---))).))))). ( -34.80) >DroYak_CAF1 102262 98 - 1 UUGUCAAGCUCUCGAGCUCACCU---GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGU--GAGCUGAAGUCACCUCGA-----GCGCA---GAGAGAGAGG ........(((((.(((((((((---(((..(((..((((.((....))....)))).))))))))))--)))))........(((..-----.....---))).))))). ( -38.70) >DroPer_CAF1 103082 100 - 1 UUGUCAAGCUCUCGGUGUCGCCUCUGAAGCUAAACAUUGCGGCAAAUGCCAAUGCAAAUGCCUCAGGG----GCUG------CCUUUAGGAGAGCUUCAUAGAGAGCC-CC ((((.((((((((((.((.((((((((.((........))((((..(((....)))..))))))))))----)).)------)))....)))))))).))))......-.. ( -38.50) >consensus UUGUCAAGCUCUCGAGCUCACCU___GAGCCGCACAUUGCCGCAAAUGCCAAUGCAAAUGCCUCAGGU__GAGCUGAAGUCACCUCGA_____GCCAACU_GAGAGUUAGC .......((((((.(((((((((...(((..(((..((((.((....))....)))).))))))))))..)))))((.......))...............)))))).... (-15.18 = -16.93 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:53 2006